A Beginner’s Guide to Molecular Docking
by James H. Zothantluanga ★ , Dipak Chetia
Academic editor: Ernest Domanaanmwi Ganaa
Sciences of Phytochemistry 1(2): 90-93 (2022); https://doi.org/10.58920/sciphy01020037
This article is licensed under the Creative Commons Attribution (CC BY) 4.0 International License.
24 Nov 2022
01 Dec 2022
02 Dec 2022
02 Dec 2022
Abstract: In this opinion, the basics of molecular docking (MD) such as binding affinity, binding pose, and ligand interactions with common docking-related terminologies (Apo protein, positive control, native ligand, co-crystal inhibitors) are discussed. We have provided different figures to aid in the graphical interpretation of the discussed literature. Following this, a few advantages (simplicity, fast, applicability) and disadvantages of MD are highlighted. This opinion will benefit bachelor and master students (or anyone) that are interested in learning the technique of MD. We encourage the sensible use of the MD technique and strict analysis to avoid interpretation errors in the results. The binding affinity, binding pose, and ligand interactions should be collectively considered during the result analysis. For every study, we strongly recommend a strict validation of the docking protocols.
Keywords: Molecular dockingBinding affinityBinding poseMolecular interactions
Introduction
Molecular docking (MD) is one of the computer-aided drug design (CADD) techniques (1). Under CADD, MD is considered a structure-based drug design (SBDD) technique (2). In the absence of a protein, SBDD such as MD cannot be carried out (3, 4). In this opinion paper, we will discuss some basics along with a few advantages and disadvantages of MD. This opinion will benefit bachelor and master students (or anyone) that are interested in learning the technique of MD.
The Basics
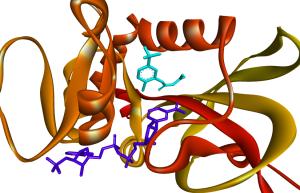
The binding affinity of a ligand towards the active binding site (orthosteric site) of a protein is studied with MD (see Figure 1A). The binding affinity of a ligand towards the non-active site (allosteric site) is also studied. Binding affinity is used synonymously with binding energy, but they have different meanings. The lower the binding energy, the higher the binding affinity, and vice versa. The unit ‘kcal/mol’ is usually associated with binding energy. Sometimes, researchers prefer to use the term ‘docking score’ instead of binding affinity/energy. Some software/web tool generates a unit-less value which is considered as docking score. Ultimately, a compound should have low binding energy, preferably lower than the native ligand (5, 6).
Native ligand (also referred to as standard/positive control/co-crystal inhibitor) is the compound that is complexed with the protein and they are usually present at the active binding site of the protein. However, not all molecules present at the active binding site are native ligands (see Figure 1B). A native ligand of a protein usually has supporting wet lab data (IC50, EC50, etc.) against the protein and this information can be found on the protein databank website. In MD, the native ligand is a positive control that serves as the benchmark with the efficacy of the test ligands will be judged. In the absence of a native ligand, it is important to search for a positive control from existing literature. The protein-free state (without any ligand) is called the Apo protein (7, 8).
The binding pose of a compound i.e. the spatial arrangement of a compound at the active binding site of a protein is studied with MD (Figure 1C). The active binding site contains all the important amino acids of the protein. One compound can have a different binding pose (spatial arrangement) at the active binding site. For instance, the PyRx virtual screening tool uses AutoDock Vina to generate 9 different binding poses for a single ligand and all the different binding poses have different binding affinities (Figure 1D) (9). A binding pose with the lowest binding energy (highest binding affinity) is usually considered the best pose. The binding pose of a compound will determine what amino acids it interacts with (10, 11).
The molecular interactions between a protein and a ligand are studied with MD. In MD (Figure 1E), it is important to make sure that a potential lead compound has interacted with the amino acids that are present at the active binding site. Sometimes, the best binding pose with a very low binding energy can have fewer molecular interactions with the amino acids present at the active binding site. Sometimes, a compound may interact with multiple amino acids except for the amino acids present at the active binding site. Sometimes, a compound may also form different types of hydrophobic/electrostatic interactions except for conventional hydrogen bonds. Preferably, a compound must form more conventional hydrogen bonds with multiple amino acids at the active site. It may be noted that a higher number of conventional hydrogen bonds correlates to a lower possibility for the development of drug resistance (12, 13).

Figure 1. (A) Red sphere indicates the active binding site (allosteric site) of SARS-CoV-2 Mpro. The native ligand bound to the protein at the active binding site is presented in yellow color. (B) Two molecules are bound to the active binding site of PfDHFR-TS. The native ligand is depicted in light blue color while the other ligand in dark blue color is a peptide. (C) The binding pose of a ligand (yellow color) within the active binding pocket of SARS-CoV-2 Mpro. (D) The 9 different binding poses for a single compound at the active binding site of PfDHFR-TS. Each binding pose represented in different colors has a different binding affinity towards the protein. (E) Protein-ligand interactions between the native ligand and the amino acids at the active binding site of SARS-CoV-2 Mpro. There are 6 different types of interaction and the bond lengths are measured in Å units.
Advantages
- In comparison to other in-silico techniques, MD is a simple technique as it can be easily and quickly executed.
- A decent laptop (a simple workstation) is all it takes to perform MD. However, the workstation is also a factor in determining the speed of MD.
- Using MD, we can study how the presence or absence of certain functional groups will impact the binding affinity of a ligand.
- MD enables to rapidly evaluate the in-silico potency of virtually designed compounds against a target protein. This is useful to identify potential leads in a drug discovery program.
- MD can be used to identify the key functional groups of a compound that will impact its binding affinity and interaction profiles. This information can be used to modify the structure of compounds to potentially improve the biological activity.
Disadvantages
- The results of MD are preliminary as they are not fully reliable to make robust conclusions.
- The results and conclusions of almost all studies carried out with MD include the term ‘potential’ as it is impossible to draw solid conclusions from the results of MD.
- The results of MD are limited as it fails to represent some real biological phenomena.
- For example, will the binding affinity remain at the same value if the docking was to run for an extended time?
- If we prolong the period of interaction, will the ligand remain stable (low binding energy) at the active site, or if it becomes unstable, what will be its fate?
- Will the ligand maintain an adequate interaction with the active amino acids if the simulation time was extended?
- Also, MD does not consider the temperature and pressure.
- The docking protocols and docking results need to be validated by carrying out additional studies. Based on our personal publishing experience, MD studies require validation with either in-vitro studies or molecular dynamics simulations.
Author Perspective
We encourage the sensible use of the MD followed by strict analysis to avoid interpretation errors in the results. The binding affinity, binding pose, and ligand interactions should be collectively considered during the result analysis. For every study, we strongly recommend a strict validation of the docking protocols. For our studies, we routinely use the academic version of PyRx virtual screening tool (https://pyrx.sourceforge.io/). We also recommend the use of DockFlin (https://etflin.com/news/2), which is a product of ETFLIN. DockFlin is a multi-ligand and a multi-protein docking tool.
Declarations
Acknowledgment
James H. Zothantluanga gratefully acknowledges the University Grant Commission and the Ministry of Tribal Affairs, Government of India for providing a UGC-SRF fellowship (Award No.: 202122-NFST-MIZ-03095) to support his Ph.D. research project. James H. Zothantluanga is also thankful to Dr. Hannah S. Lalnunpuii for providing all the necessary help during manuscript preparation.
Ethics Statement
Not applicable.
Data Availability
Not applicable.
Funding Information
Not applicable.
Conflict of Interest
The authors declare no conflicting interest.
References
- Zothantluanga JH (2021) Molecular Docking Simulation Studies, Toxicity Study, Bioactivity Prediction, and Structure-Activity Relationship Reveals Rutin as a Potential Inhibitor of SARS-CoV-2 3CL pro. J Sci Res 65:96–104.
- Umar AK, Zothantluanga JH (2021) Structure-Based Virtual Screening and Molecular Dynamics of Quercetin and Its Natural Derivatives as Potent Oxidative Stress Modulators in ROS-induced Cancer. Indones J Pharm 3:60.
- Umar AK, Kelutur FJ, Zothantluanga JH (2021) Flavonoid Compounds of Buah Merah (Pandanus conoideus Lamk) as a Potent Oxidative Stress Modulator in ROS-induced Cancer: In Silico Approach. Maj Obat Tradis 26:221.
- Pasala PK, Uppara RK, Rudrapal M, Zothantluanga JH, Umar AK (2022) Silybin phytosome attenuates cerebral ischemia‐reperfusion injury in rats by suppressing oxidative stress and reducing inflammatory response: In vivo and in silico approaches. J Biochem Mol Toxicol. 36:e23073.
- Zothantluanga JH, Gogoi N, Shakya A, Chetia D, Lalthanzara H (2021) Computational guided identification of potential leads from Acacia pennata (L.) Willd. as inhibitors for cellular entry and viral replication of SARS-CoV-2. Futur J Pharm Sci 7:201.
- Pasala PK, Abbas Shaik R, Rudrapal M, Khan J, Alaidarous MA, Jagdish Khairnar S, et al. (2022) Cerebroprotective effect of Aloe Emodin: In silico and in vivo studies. Saudi J Biol Sci 29:998–1005.
- Zothantluanga J, Aswin SK, Rudrapal M, Chetia D (2021) Antimalarial Flavonoid-Glycoside from Acacia pennata with Inhibitory Potential Against PfDHFR-TS: An In-silico Study. Biointerface Res Appl Chem 12:4871–87.
- Patowary L, Borthakur MS, Zothantluanga JH, Chetia D (2021) Repurposing of FDA approved drugs having structural similarity to artemisinin against PfDHFR-TS through molecular docking and molecular dynamics simulation studies. Curr Trends Pharm Res 8:14–34.
- Dallakyan S, Olson AJ (2015) Small-Molecule Library Screening by Docking with PyRx. Methods Mol Biol 1263:243–50.
- Zothantluanga JH, Abdalla M, Rudrapal M, Tian Q, Chetia D, Li J (2022) Computational Investigations for Identification of Bioactive Molecules from Baccaurea ramiflora and Bergenia ciliata as Inhibitors of SARS-CoV-2 M pro. Polycycl Aromat Compd. https://doi.org/10.1080/10406638.2022.2046613.
- Rudrapal M, Celik I, Khan J, Ansari MA, Alomary MN, Yadav R, et al. (2022) Identification of bioactive molecules from Triphala (Ayurvedic herbal formulation) as potential inhibitors of SARS-CoV-2 main protease (Mpro) through computational investigations. J King Saud Univ - Sci 34:101826.
- Umar AK, Zothantluanga JH, Aswin K, Maulana S, Sulaiman Zubair M, Lalhlenmawia H, et al. (2022) Antiviral phytocompounds “ellagic acid” and “(+)-sesamin” of Bridelia retusa identified as potential inhibitors of SARS-CoV-2 3CL pro using extensive molecular docking, molecular dynamics simulation studies, binding free energy calculations, and bioactivi. Struct Chem. 33:1445–65.
- Rudrapal M, Celik I, Chinnam S, Azam Ansari M, Khan J, Alghamdi S, et al. (2022) Phytocompounds as potential inhibitors of SARS-CoV-2 Mpro and PLpro through computational studies. Saudi J Biol Sci. 29:3456–65.
 ETFLIN
Notification
ETFLIN
Notification