Characterization, In Silico Antimalarial, Antiinflammatory, Antioxidant, and ADMET Assessment of Neonauclea excelsa Merr.
by Neksumi Musa , Mubarak Muhammad Dahiru , Enoch Buba Badgal ★
Academic editor: Adeleye Ademola Olutayo
Sciences of Pharmacy 3(2): 92-107 (2024); https://doi.org/10.58920/sciphar0302232
This article is licensed under the Creative Commons Attribution (CC BY) 4.0 International License.
19 Mar 2024
01 May 2024
04 May 2024
08 May 2024
Abstract: In our study, we identified the phytoconstituents and carried out antimalarial, anti-inflammatory, antioxidant, and ADMET assessments of Neonauclea excelsa. The phytochemicals were detected and quantified followed by identification via GC-MS. The antimalaria, anti-inflammatory, and antioxidant assessments were done by molecular docking (MD) and molecular dynamics simulation (MDS) while ADMET by ADMET predictions. Saponins (27.33% ±1.20) and terpenes (8.33% ±0.73) were detected while alkaloids, steroids, glycosides, and flavonoids were absent. Exactly 29 compounds were identified with squalene being the most abundant (32.41%). Compound II exhibited the lowest BA (-6.4 kcal/mol) and Ki (20.12 µM), interacting with dihydrofolate reductase-thymidylate synthase. IV exhibited the lowest respective BA and Ki interacting with Plasmodium falciparum hexose transporter protein 1 (-6.2 kcal/mol and 28.20 µM), cyclo-oxygenase-2 (-7.2 kcal/mol and 5.21 µM), and myeloperoxidase (-7.4 kcal/mol and 3.71 µM). Compound VII had the lowest respective BA and Ki interacting with inducible nitric oxide synthase (-8.0 kcal/mol and 1.35 µM), xanthine oxidase (-7.2 kcal/mol and 5.21 µM), and cytochrome p450 21A2 (-7.0 kcal/mol and 7.30 µM). The MDS showed various cluster mobilities and residue fluctuations up to 5.26, 2.96, 5.10, 3.51, 5.02, 4.65, and 6.18 Å for dihydrofolate reductase-thymidylate synthase, Plasmodium falciparum hexose transporter protein 1, inducible nitric oxide synthase (INOS), cyclo-oxygenase-2 (COX2), xanthine oxidase (XO), cytochrome p450 21A2, and myeloperoxidase, respectively. Additionally, these compounds demonstrated good pharmacological properties with minimal toxicity. Conclusively, the identified compounds might be significant contributors to the antimalarial, anti-inflammatory, and antioxidant activity of N. excelsa and are good sources of novel antimalarial, anti-inflammatory, and antioxidant drugs.
Keywords: GCMSInflammationMalariaMolecular dockingMolecular dynamicsOxidative stress
Introduction
Malaria is an ailment transmitted via a bite by an infected female Anopheles mosquito transferring the Plasmodium parasite to an individual. These parasites include 5 species; Plasmodium falciparum, P. vivax, P. malariae, P. ovale, and P. knowlesi. However, the P. falciparum and P. vivax are the most menacing with the former being the deathliest and most prevalent of all the parasites in Africa (1). Malaria continues to exert devastating effects worldwide despite several previous and continued interventions by the World Health Organization (WHO). A 2022 epidemiological data shows 608,000 malaria deaths out of 249 million cases across 85 countries with Africa at the leading position globally (1-3). Moreover, the highest prevalence (94%) and number of deaths (95%) reported in 2022 were in Africa with children being up to 80% of the deaths (1-3). Furthermore, Nigeria tops the list of malaria deaths from Africa with 26.8% next to the DR Congo, Uganda, and Mozambique with 12.3%, 5.1%, and 4.2%, respectively (1-3). There are different treatment options for malaria, including the combined artemisinin-based therapy, chloroquine, and primaquine targeting falciparum, vivax, and ovale, respectively (1, 4, 5). However, the emergence of resistance poses further threats to treatments. Plant-based treatments offer an affordable and available alternative anti-malarial therapy targeting multiple proteins required for the parasite's survival.
Medicinal plants have been finding their way into therapeutic uses in traditional medicine due to their proven efficacy against different ailments. Different parts of medicinal plants made up of roots bark and leaves are prepared in powder or decoction and are taken by oral, topical, or inhalation in different doses to achieve relief from ailments (6). In Nigeria, different preparations in the form of decoctions and infusions of different plant parts are employed in the treatment of different ailments (7). The biological activities of plants are attributed to their phytochemical composition made up of different bioactive compounds. The extensive application of plant-based drugs in the prevention or fighting of several diseases is due to their phytoconstituents (8-11). Some of these phytochemicals include alkaloids, saponins, glycosides, and flavonoids. The antioxidant activities of some of the plants were previously reported (12-14). Neonauclea excelsa plant is a shrub from the Rubiaceae family found in tropical Africa. The plant is made of fruits, which have a lot of brownish seeds embedded within it. Several studies have reported the biological activities of N. excelsa which include antioxidant and antidiabetic activities (15). Furthermore, the anti-inflammatory (16) and anti-plasmodial (17) activities of N. excelsa were previously reported.
Inflammation and oxidative stress are closely associated with the pathogenesis of malaria infection (18-22). The antimalarial response to the Plasmodium parasite includes an increased reactive oxygen species (ROS) production by the host phagocytes with increased antioxidant enzyme activities including xanthine oxidase (19, 20, 23). This inherent response specifically targets the parasite environment, causing damage to the parasite. However, persistent ROS generation leading to oxidative stress might subsequently trigger the inflammatory cascades exacerbating the condition by causing severe tissue damage to the host (23). Moreover, the continued inflammatory cascades further aggravate the response to the ROS generation (20). Furthermore, the continued hemoglobin depletion by the parasite in the red blood cells (RBC) liberates the heme, further exacerbating the host's oxidative stress and complicating the ailment (24). Thus, therapies targeting oxidative stress and inflammation including plant-based drugs might play a vital role in malarial treatments, suppressing the severity of these responses and minimizing host impairment. In our previous study, we reported some of these anti-inflammatory and antioxidant targets, depicting their interactions with different compounds via molecular docking and molecular dynamics simulations (25). In the present study, we explore the phytochemistry of N. excelsa via GC-MS characterization and the potential application of its phytoconstituents in malaria therapy, using molecular docking, molecular dynamics simulations, and ADMET prediction.
Experimental Section
Materials
Plant Material
N. excelsa was collected from Girei Local Government Area (9.3581º N and 12.5430º E) of Adamawa state, Nigeria in the month of June, 2022. This was followed by its identification and authentication by a Forest Technologist from the Forestry Technology Department of Adamawa State Polytechnic Yola. A voucher specimen (no. ASP/FT/0092) was deposited in the departmental herbarium. The stem bark was air-dried and ground into powder with a blender.
Chemicals and Reagents
Methanol, chloroform, ethyl acetate, butanol, and diethyl ether were purchased from Xingtai Dakun Technology Co., Ltd (China). All the chemicals and reagents employed in the present study were of analytical grade.
Methods
N. excelsa bark powder (300 g) was extracted by maceration with 1 L of 70% (v/v) ethanol for 2 days at room temperature with agitation every 24 hours. This was followed by filtering the extract using Whatman no.1 filter paper and subsequently concentrated to dryness under reduced pressure at 40 ºC (26).
Phytochemical Analysis
The phytochemicals present were identified using the method described by Evans (26) to check for the presence of alkaloids, saponins, steroids, glycosides, terpenoids, and flavonoids.
Determination
of Total Saponins
Total saponins were quantified by the method previously described (27). Exactly 0.2 g extract was introduced into a conical flask and 10 mL of 20% aqueous ethanol was added followed by heating in a water bath for 1 h at 55°C. This was transferred into a 250 mL separator funnel and 5 mL of diethyl ether was added and shaken vigorously. After complete separation, the ether layer was discarded while the aqueous layer was recovered. Exactly 10 mL of n-butanol and 2 mL of 5% aqueous NaCl were added. The upper layer formed was recovered and dried to a constant weight in an oven. The total saponins were calculated according to Equation 1.
Equation 1
Equation 2
Where T = 298.15 K (temperature), R = 1.985 x 10-3 kcal-1.mol-1.k-1 (the universal gas constant), ∆G = BA.
Table 1. Screened Compounds for the MD.
|
No. |
Name |
Designation |
PubChem ID |
|
1 |
Catechol |
I |
289 |
|
2 |
Buminafos |
II |
39966 |
|
3 |
4-Methylanisole |
III |
7731 |
|
4 |
Methyl 6-morpholinonicotinate |
IV |
2776466 |
|
5 |
1-(4-bromobutyl) piperidin-2-one |
V |
536377 |
|
6 |
1-Chlorooctadecane |
VI |
18815 |
|
7 |
Farnesol |
VII |
445070 |
Determination of Total Terpenoids
Total terpenoids were quantified by the gravimetric method described previously (28). Exactly 0.2 g of the extract was soaked in 10 mL of ethanol for 24 hours., followed by filtration and extraction with 10 mL of petroleum ether using a separating funnel. The ether layer was collected in a pre-weighed crucible and dried in the oven. The total terpenoid content was measured according to Equation 1.
Gas Chromatography-Mass Spectrometry (GC-MS) Analysis
The compounds present in the plant were detected and identified by a Gas chromatography-mass spectrometer (7890, 5975 Agilent, USA) fitted with a fused silica column (30 X0.25 mm, with 1 µm film thickness). The procedure was as we described previously (29).
Molecular Docking (MD) and Molecular Dynamics Simulation (MDS)
The MD and MDS were carried out using a laptop PC (HP Pavillion x360 Convertible 14-dw1xx) with an 11th Gen Intel(R) Core i3-1115G4 @ 3.00GHz processor, 8.00 GB RAM, and Windows 11. The compounds identified from the GS-MS analysis were initially screened by applying Lipinski’s rule using the DruLiTo software. Only compounds that passed Lipinski’s rule in addition to either the Ghose filter or Veber filter were selected for the MD. The compounds were downloaded from the PubChem database in SDF format while the target proteins were from the RSCB database. These proteins were selected due to their involvement in the pathology of malaria, inflammation, and oxidative stress. The compounds including the PubChem ID are presented in Table 1 while the docking targets along with their grid box in Table 2. The docking pockets (grid box) within the targets were identified using the PrankWeb online server.
The MD was done via the vina interface of the PyRx software to determine the binding energy (BA) and binding interactions of the docked complexes. Initial energy minimization of the compounds was carried out using the PyRx software while the targets were prepared using the AutoDockTools (v 1.5.7) (30) by removing identical chains, heteroatoms, and water molecules, leaving only the apoprotein. Furthermore, the exhaustiveness of the docking was set to 16. Finally, the 2D and 3D compound-protein docked complex was visualized using LigPlot+ (version 2.2.8) and PyMOL (version 2.5.4) software, respectively. The former was used to visualize the hydrogen bond (HB) and hydrophobic (HPB) interactions while other interactions were with the protein-ligand interaction profiler webserver (31). The inhibition constant (Ki) was evaluated from Equation 2. Additionally, only the top-docked compounds were further subjected to the MDS. Moreover, the MDS was done via CABs-flex v2.0 (32) and WebNMA (33) online servers to determine the root-mean-square fluctuations and cluster mobility of the docked complexes, respectively.
Absorption, Distribution, Metabolism,
Excretion, and Toxicity (ADMET) Prediction
The ADMET of the compounds with the most favorable docking interactions with the protein targets were predicted using the pkCSM online server (34) while drug likeliness was determined with the SwissADME online server (35).
Table 2. Anti-malarial, antiinflammatory, and antioxidant protein targets.
|
Activity |
Targets |
PDB ID |
Grid Box |
||
|
X |
Y |
Z |
|||
|
Antimalaria |
Dihydrofolate reductase-thymidylate synthase (DHFRTS) |
2BL9 |
89.98 |
9.29 |
31.17 |
|
Plasmodium falciparum hexose transporter (PfHT1) |
6M2L |
-23.84 |
-46.98 |
-0.12 |
|
|
Antiinflammatory |
Inducible nitric oxide synthase (INOS) |
4NOS |
0.55 |
96.89 |
19.64 |
|
Cyclo-oxygenase-2 (COX2) |
5IKT |
165.63 |
186.01 |
193.83 |
|
|
Anti-oxidant |
Xanthine Oxidase (XO) |
3NVZ |
23.23 |
-16.04 |
35.18 |
|
Cytochrome P450 21A2 (CytP450 21A2) |
4Y8W |
-14.66 |
11.96 |
28.61 |
|
|
Myeloperoxidase (MPO) |
7LAL |
19.01 |
-13.64 |
-4.48 |
|
Statistical Analysis
The values are expressed as mean ± standard error. Statistical Package for the Social Sciences (SPSS) version 22 Software was used for the statistical analysis.
Figure 1. Structures of identified compounds in ethanol extract of Neonauclea excelsa.
Table 3. Phytochemical composition of ethanol extract of Neonauclea excelsa.
|
No. |
Phytochemical |
Inference |
Concentration (%) |
|
1 |
Alkaloids |
- |
- |
|
2 |
Saponins |
+ |
27.33 ±1.20 |
|
3 |
Steroids |
- |
- |
|
4 |
Glycosides |
- |
- |
|
5 |
Terpenoids |
+ |
8.33 ±0.73 |
Note: (+) = present and (-) = absent. Concentration values are in triplicate determinations (± SEM).
Table 4. Compounds identified in Neonauclea excelsa.
|
S/N |
Name of compound |
Retention Time |
Peak Area (%) |
Molecular weight |
Formula |
|
1 |
Catechol |
4.060 |
2.19 |
110.11244 |
C6H6O2 |
|
2 |
n-Hexadecane |
4.523 |
1.16 |
226.44596 |
C16H34 |
|
3 |
Buminafos |
4.792 |
0.55 |
347.47838 |
C18H38NO3P |
|
4 |
n-Undecane |
5.187 |
0.43 |
156.31156 |
C11H24 |
|
5 |
Citronellic acid |
5.479 |
0.38 |
170.25172 |
C10H18O2 |
|
6 |
Octadecane |
5.914 |
1.25 |
254.49972 |
C18H38 |
|
7 |
4-Methylanisole |
6.217 |
0.22 |
122.1668 |
C8H10O |
|
8 |
Methyl 6-morpholinonicotinate |
6.692 |
0.37 |
222.24376 |
C11H14N2O3 |
|
9 |
13-Tetradecenal |
6.841 |
0.35 |
210.198365 |
C14H26O |
|
10 |
Methyl palmitate |
6.984 |
2.10 |
270.45576 |
C17H34O2 |
|
11 |
Ethyl tridecanoate |
7.499 |
5.27 |
242.402 |
C15H30O2 |
|
12 |
Methyl oleate |
8.437 |
3.25 |
296.49364 |
C19H36O2 |
|
13 |
Methyl 10-methyldodecanoate |
8.620 |
2.25 |
228.37512 |
C14H28O2 |
|
14 |
(9Z)-octadeca-9,17-dienal |
8.923 |
6.38 |
264.45148 |
C18H32O |
|
15 |
Ethyl stearate |
9.141 |
5.56 |
312.5364 |
C20H40O2 |
|
16 |
Palmitic acid |
9.621 |
3.05 |
256.42888 |
C16H32O2 |
|
17 |
1-(4-bromobutyl) piperidin-2-one |
9.976 |
4.79 |
234.13614 |
C9H16BrNO |
|
18 |
(Z,Z)-7,11-Hexadecadien-1-ol, acetate |
10.314 |
2.66 |
280.45088 |
C18H32O2 |
|
19 |
cis-Vaccenic acid |
10.577 |
1.05 |
282.46676 |
C18H34O2 |
|
20 |
tert-Hexadecyl mercaptan |
10.760 |
2.70 |
258.50596 |
C16H34S |
|
21 |
2-Methyloctadeca-3,13-dien-1-ol |
11.138 |
1.88 |
280.49424 |
C19H36O |
|
22 |
1-Chlorooctadecane |
11.550 |
1.62 |
288.94478 |
C18H37Cl |
|
23 |
(E)-5-Icosene |
11.933 |
1.86 |
280.5376 |
C20H40 |
|
24 |
stearyloxyethylene |
12.322 |
0.74 |
296.537 |
C20H40O |
|
25 |
Squalene |
13.884 |
32.41 |
410.727 |
C30H50 |
|
26 |
Tridecyloxirane |
14.519 |
2.88 |
226.4026 |
C15H30O |
|
27 |
(Z)-9-Tetradecenal |
14.874 |
4.58 |
210.35984 |
C14H26O |
|
28 |
3-(2',2'-dimethyl-6'-methylidenecyclohexyl)-1-propanol |
15.143 |
0.99 |
182.30 |
C12H22O |
|
29 |
Farnesol |
15.378 |
2.93 |
222.37084 |
C15H26O |
Result
Phytochemical Analysis
The phytochemicals detected and their concentration are shown in Table 3. Saponins and terpenoids were detected while alkaloids, steroids, glycosides, and flavonoids were absent. Saponins were quantified up to 27.33% ±1.20 while terpenoids were in the concentration of 8.33% ±0.73.
GC-MS Analysis
The various compounds identified with their retention time, peak area, molecular weight, and formula are depicted in Table 4. GC-MS analysis revealed 29 compounds liable for the various pharmacological effects activities of the plant. Squalene had a peak area of 32.41% which is the highest, followed by (9Z)-octadeca-9,17-dienal with a peak area of 6.38%, then 1-(4-bromobutyl) piperidin-2-one (4.79%), and (Z)-9-Tetradecenal (4.58%).
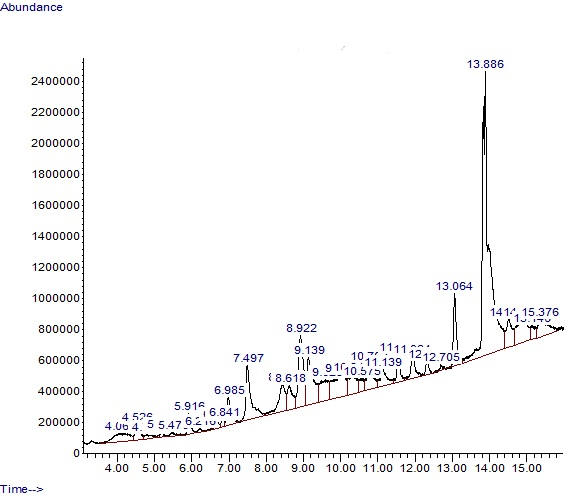
Figure 2. GC-MS Chromatogram of compounds identified in ethanol extract of Neonauclea excelsa.
The structures of the different compounds identified and the chromatogram of the GC-MS analysis showing the different peaks at different retention times are presented in Figures 1 and 2, respectively. The majority of the identified compounds were long-chain fatty acids including Methyl palmitate, palmitic acid, and vaccenic acid. Other compounds include those containing aromatic rings.
Molecular Docking and Dynamics Simulation
The docking interactions of the top-docked compounds with the antimalarial, anti-inflammatory, and antioxidant targets are presented in Table 5 revealing the BA, Ki, and binding interactions. Compound II demonstrated the most favorable docking interaction with DHFRTS, exhibiting the lowest BA (-6.4 kcal/mol) and Ki (20.12 µM), in addition to 2 HBs and 11 HPBs while IV exhibited a slightly lower BA (-6.3 kcal/mol), Ki (23.82 µM), and HPBs (8) though with higher HBs (3). Although both IV and II exhibited the same BA (-6.2 kcal/mol) and Ki (28.20 µM) while interacting with PfHT1, IV had a higher number of HPBs (12), in addition to a salt bridge formation with Lys51.
The docking interaction with the anti-inflammatory targets shows a favorable interaction of VII with INOS, exhibiting the lowest BA (-8 kcal/mol) and Ki (1.35 µM), next to II with BA and Ki of -7.3 kcal/mol and 4.40 µM, respectively. Moreover, VII had an HB, absent in II though it has higher HPBs (15). Compound IV exhibited the lowest BA (-7.2 kcal/mol) and Ki (5.21 µM), leading to a favorable docking interaction with COX2 than VII. Although IV exhibited the most favorable docking interaction in addition to a salt bridge with Arg120, VII had higher HBs (2) and HBPs (14).
Table 5. Docking interactions of the compounds with the antimalarial, anti-inflammatory, and antioxidant targets.
|
Activity |
Targets |
Compounds |
BA (kcal/mol) |
Ki (µM) |
HBs |
HPBs |
Salt Bridge |
|
Antimalarial |
DHFRTS |
II |
-6.4 |
20.12 |
2 |
11 |
- |
|
IV |
-6.3 |
23.82 |
3 |
8 |
- |
||
|
PfHT1 |
IV |
-6.2 |
28.20 |
1 |
12 |
Lys51 |
|
|
II |
-6.2 |
28.20 |
2 |
6 |
- |
||
|
Anti-inflammatory |
INOS |
VII |
-8 |
1.35 |
1 |
11 |
- |
|
II |
-7.3 |
4.40 |
- |
15 |
- |
||
|
COX2 |
IV |
-7.2 |
5.21 |
- |
11 |
Arg120 |
|
|
VII |
-7 |
7.30 |
2 |
14 |
|
||
|
Antioxidant |
XO |
VII |
-7.2 |
5.21 |
2 |
12 |
- |
|
IV |
-7.1 |
6.16 |
4 |
5 |
- |
||
|
CytP450 21A2 |
VII |
-7 |
7.30 |
4 |
7 |
- |
|
|
IV |
-6.8 |
10.23 |
2 |
10 |
His366 |
||
|
MPO |
IV |
-7.4 |
3.71 |
1 |
9 |
|
|
|
VII |
-6.6 |
14.35 |
1 |
13 |
|
The lowest BA (-7.2 kcal/mol) and Ki (5.21 µM) were exhibited by VII among the compounds, interacting with XO with higher HPBs (12) than IV. However, IV had higher HBs (4). Furthermore, this compound (VII) demonstrated the lowest BA (-7 kcal/mol) and Ki (7.30 µM), exhibiting the most favorable interaction with CytP450 21A2. However, IV had higher HPBs (10) with an additional salt bridge formation with His366.
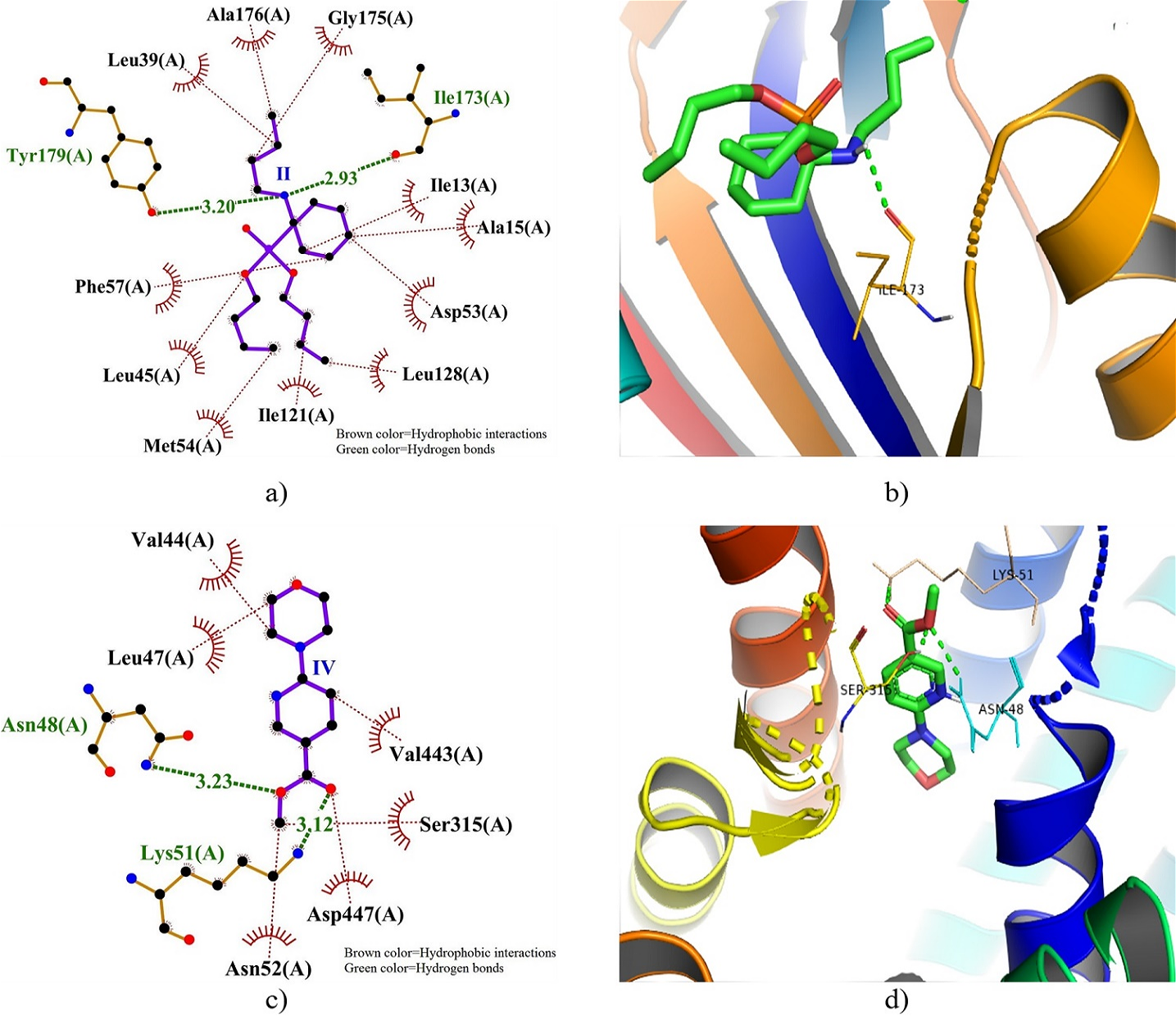
Figure 3. 2D and 3D docking interactions of DHFRTS and PfHT1 with compound II and IV. Note: a) 2D interaction of compound II with DHFRTS, b) 3D interaction of compound II with DHFRTS, c) 2D interaction of compound IV with PfHT1, and d) 3D interaction of compound IV with PfHT1.
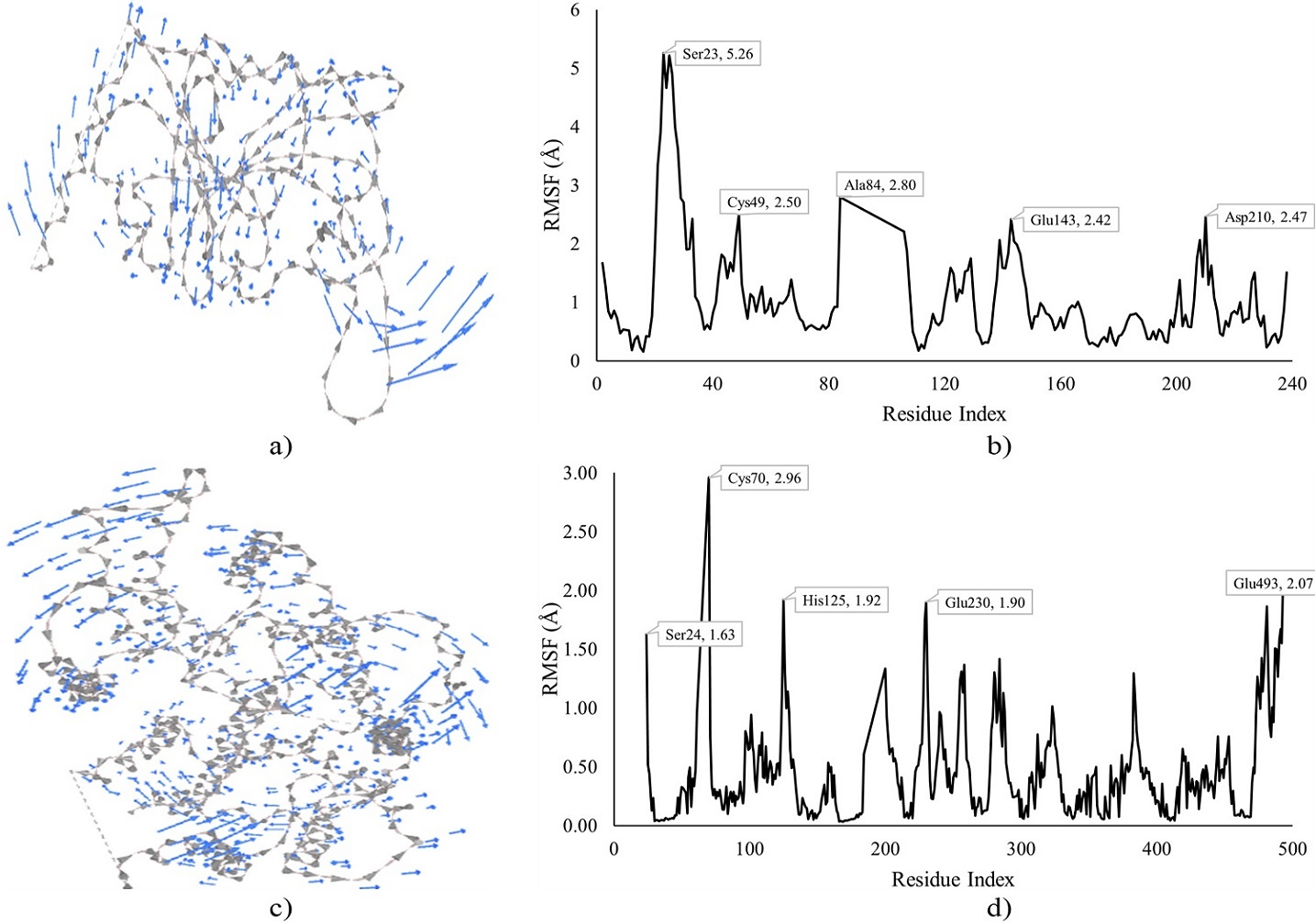
Figure 4. MDS of DHFRTS and PfHT1 docked with compound II and IV. Note: a) Cluster mobility of II with DHFRTS, b) RMSF of II with DHFRTS, c) Cluster mobility of IV with PfHT1, and d) RMSF of IV with PfHT1.
Furthermore, IV showed better docking interaction with MPO, exhibiting lower BA (-7.4 kcal) and Ki (3.71 µM) though VII had higher HPBs (13). The 2D and 3D dock poses of the interactions of DHFRTS and PfHT1 with compounds II and IV, respectively are presented in Figure 3, depicting the HB distances and HPB residues. An HB was formed by II with Tyr179 and Ile173 residues of DHFRTS with distances of 3.20 and 2.93, respectively, surrounded by HPBs.
The MDS of compound II and IV docked complexes of DHFRTS and PfHT1, respectively is depicted in Figure 4, revealing the cluster mobility and residue fluctuations. Higher residue fluctuations and cluster mobility were observed at the N-terminals of the protein for both DHFRTS and PfHT1. The highest fluctuation for the II-DHFRTS docked complex was observed at Ser23 (5.26 Å) next to Ala84 (2.80 Å), Cys49 (2.50 Å), Asp210 (2.47 Å), and Glu43 (2.42 Å). Cys70 (2.96 Å) was the most fluctuated for the IV-PfHT1 docked complex, followed by Glu493 (2.07 Å) at the C-terminal of the protein. Other fluctuated residues for the IV-PfHT1 complex include His125 (1.92 Å), Glu230 (1.90 Å), and Ser24 (1.63 Å).
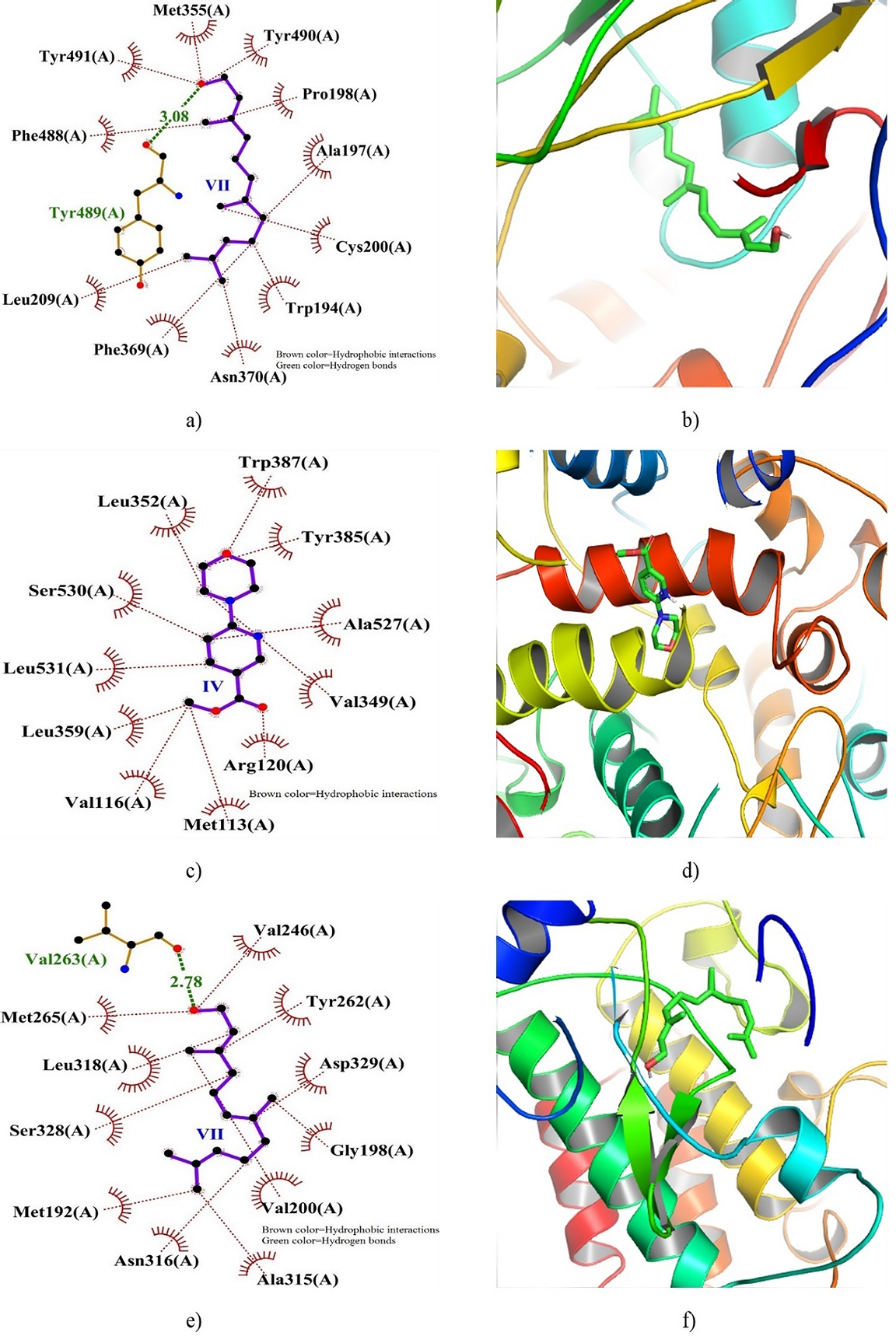
Figure 5. 2D and 3D docking interactions of INOS, COX2, and IRAK4 with compound VII, IV, and VII. Note: a) 2D interaction of VII with INOS, b) 3D interaction of VII with INOS, c) 2D interaction of IV with COX2, and d) 3D interaction of IV with COX2.
Figure 5 illustrates the 2D and 3D dock poses of INOS, COX2, and IRAK4 in complex with VII, IV, and VII, respectively. The depiction highlights the residues pivotal in the formation of hydrogen bonds (HB) and hydrophobic binding interactions (HPBs), along with the distances of HBs. Notably, a hydrogen bond (with a distance of 3.08 Å) was discerned between Tyr489 of INOS and VII, alongside multiple HPBs fostering stability within the binding site. Conversely, while COX2 interaction with IV lacked any observed HB, numerous HPBs contributed to the stabilization of the docked complex. Furthermore, the interaction of IRAK4 with VII revealed the formation of an HB (2.78 Å) with Val263 situated within a hydrophobic pocket, encircled by several HPB interactions.
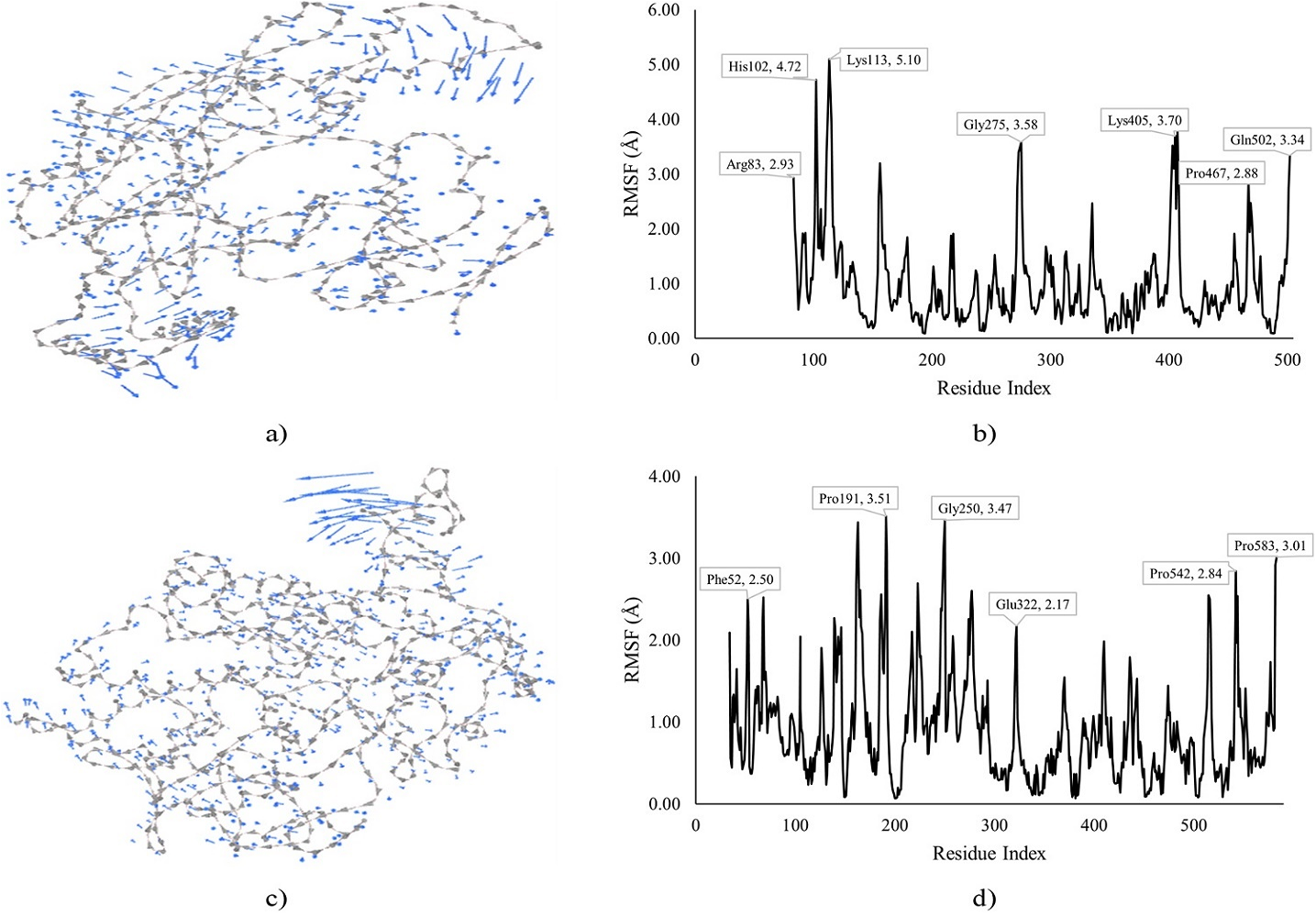
Figure 6. MDS of INOS and COX2 docked with compound VII and IV. Note: a) Cluster mobility of VII with INOS, b) RMSF of VII with INOS, c) Cluster mobility of IV with COX2, and d) RMSF of IV with COX2.
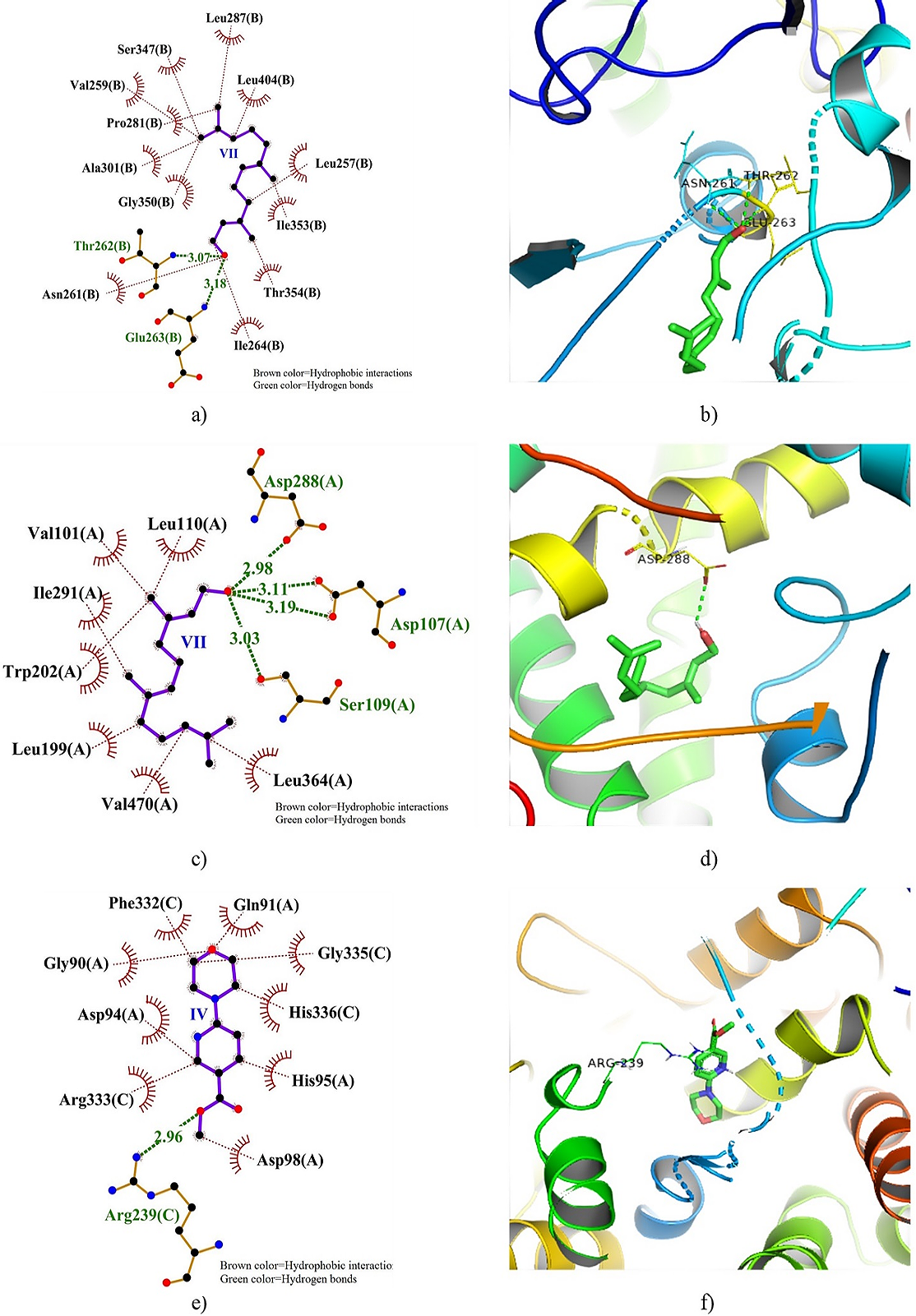
Figure 7. 2D and 3D docking interactions of XO, CytP450 21A2, and MPO with compound VII, VII, and IV. Note: a) 2D interaction of VII with XO, b) 3D interaction of VII with XO, c) 2D interaction of VII with CytP450 21A2, d) 3D interaction of IV with CytP450 21A2, e) 2D interaction of IV with MPO, and f) 3D interaction of IV with MPO.
The MDS of VII-INOS and IV-COX2 docked complexes is shown in Figure 6, depicting the cluster mobility and RMSF. The VII-INOS docked complex showed higher cluster mobility towards the N-terminal with His102 and Lys113 exhibiting RMSF of 4.72 and 5.10 Å, respectively. An RMSF of 3.58 Å was exhibited by Gly275 close to mid-chain while Lys405 and Gln502 were higher at the C-terminal of the protein with RMSF of 3.70 and 3.34 Å, respectively. The IV-COX2 docked complex exhibited cluster mobility and higher RMSF close to mid-chain with Pro191 and Gly250 being highly fluctuated up to 3.51 and 3.47 Å, respectively. Furthermore, the RMSF demonstrated by Pro543 (2.84 Å) and Pro583 (3.01 Å) at the C-terminal were higher than the RMSF of Phe52 (2.50 Å) at the N-terminal of the protein.
Figure 7 displays the 2D and 3D dock poses of XO, CytP450 21A2, and MPO docked with VII, VII, and IV, respectively, showing the residues involved in HBs and HPBs. Notable interaction of XO with VII includes HB formation with Thr262 and Glu263 with bond distances of 3.07 and 3.18 Å, respectively, surrounded by many HPBs. A total of 4 HBs was formed between VII and CytP450 21A2 with Asp107 forming two with bond distances of 3.11 and 3.19 Å. Additionally, HBs was formed with Asp288 (2.98 Å) and Ser109 (3.03 Å). The IV-MPO docked complex showed clusters of HPBs surrounding IV though a single HB was observed with Arg239 with a distance of 2.96 Å.
The MDS of XO, CytP450 21A2, and MPO docked with VII, VII, and IV, respectively is shown in Figure 8, exhibiting the cluster mobility and RMSF of the residues. The MDS of the VII-XO docked complex shows cluster mobility with the highest (5.02 Å) observed at the C-terminal by Glu143, next to Lys250 (4.22 Å). Moreover, Pro224 (3.39 Å) was the most fluctuated at the N-terminal though lower than Ala500 (4.20 Å) close to the mid-chain of the protein. A similar pattern was observed in the VII-CytP450 21A2 docked complex with higher fluctuations observed at the C-terminal by Gly414 (5.02 Å) followed by Gly456 and Gly485 with RMSF of 3.67 and 3.17 Å, respectively. Furthermore, a higher RMSF (3.53 Å) was exhibited by Gln276 at mid-chain while Gly131 was the most fluctuated (2.71 Å) towards the N-terminal. The MDS of the IV-MPO docked complex showed the highest (6.18 Å) fluctuation by Val120 towards the N-terminal, next to Cys1 (3.32 Å). Moreover, Pro269 (2.34 Å) and 352 (3.19 Å) notably fluctuated at mid-chain while Pro558 had the highest RMSF (2.93 Å) at the C-terminal.
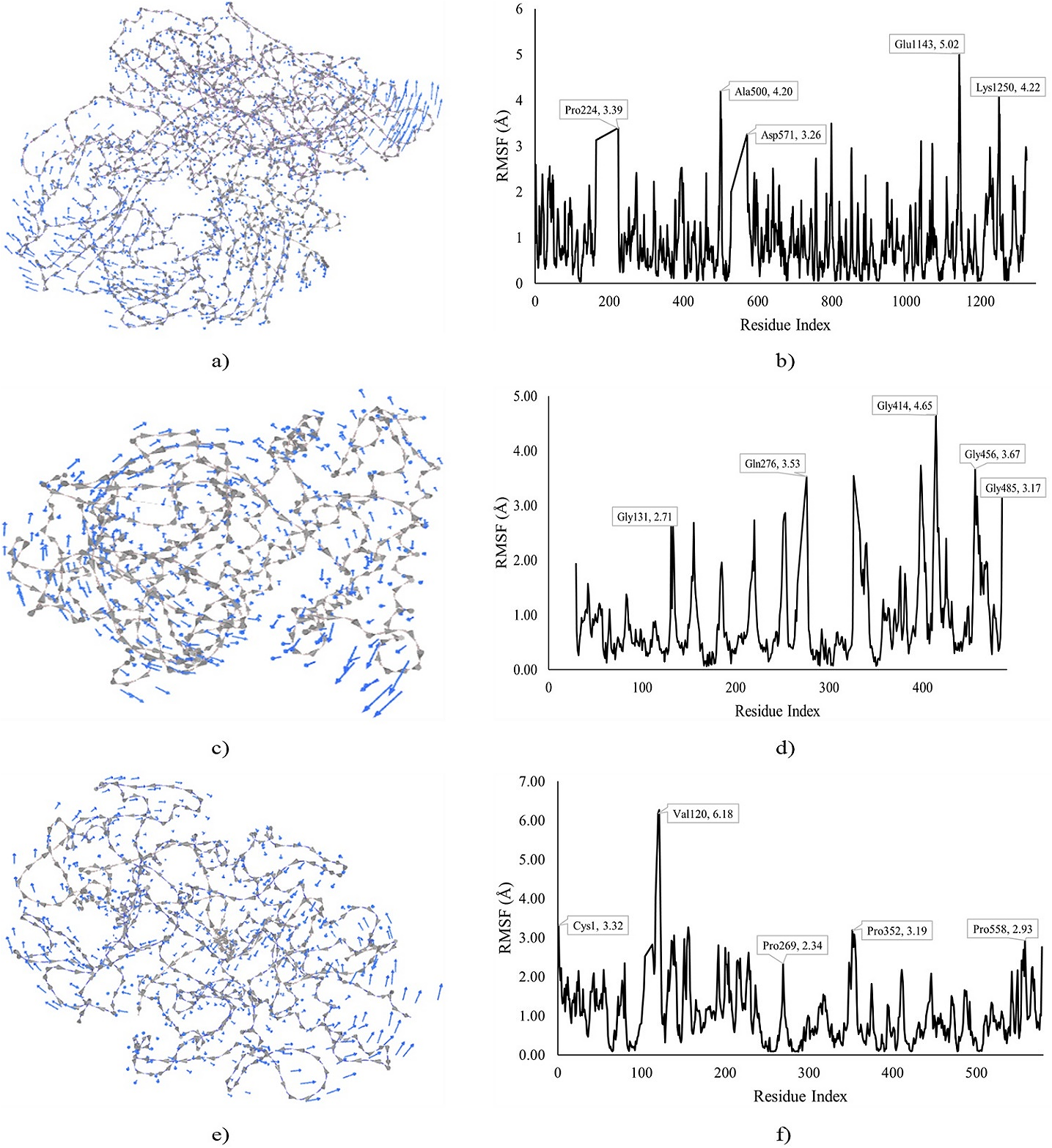
Figure 8. MDS of XO, CytP450 21A2, and MPO docked with VII, VII, and IV. Note: a) Cluster mobility of VII with XO, b) RMSF of VII with XO, c) Cluster mobility of VII with CytP450 21A2, d) RMSF of VII with CytP450 21A2, e) Cluster mobility of IV with MPO, and d) RMSF of IV with MPO.
ADMET Prediction
Table 6 presents the ADMET predictions of the top-docked compounds (II, IV, and VII), revealing their predicted pharmacokinetic properties. The highest water solubility was exhibited by IV (-1.72 log mol/L) followed by II and VII with -3.81 and -5.39 log mol/L, respectively. II showed the highest (1.70) Caco2 permeability while the lowest was exhibited by IV (1.26) though it has the highest (100%) gastrointestinal absorption next to VII (91.53%). Furthermore, IV showed the highest skin permeability (-3.18 log Kp) followed by II and VII with -2.70 and -1.51 log Kp, respectively. Additionally, all the compounds were neither substrates nor inhibitors of P-glycoprotein.
Compound VII, characterized by the highest human volume of distribution (0.36 log VDss), demonstrated notable distinctions in comparison to II (0.27 VDss) and IV (-0.38 log VDss), particularly in terms of fraction unbound where IV exhibited the highest (0.56). Moreover, the enhanced attributes of VII extended to its superior blood-brain barrier permeability (0.66 log BB) and central nervous system permeability (-1.9 log PS) compared to II and IV, further emphasizing its potential therapeutic efficacy. Notably, all compounds under scrutiny showed neither substrate nor inhibitory activity towards CYP2D6, CYP3A4, CYP1A2, CYP2C19, CYP2C9, and CYP3A4 enzymes, underlining their favorable pharmacokinetic profiles. Furthermore, among the compounds, VII stood out with the highest total clearance (1.75 log ml/min/kg), closely followed by II (0.76) and IV (0.58), showcasing their distinct metabolic pathways. Additionally, the compounds did not exhibit renal OCT2 substrate activity, adding another layer of insight into their metabolic and excretory characteristics.
Table 6. ADMET predictions of the top-docked compounds.
|
ADMET Parameters |
Compounds |
|||
|
II |
II |
II |
||
|
Absorption |
Water solubility (log mol/L) |
-3.81 |
-1.72 |
-5.39 |
|
Caco2 permeability (log Papp in 10 cm/s) |
1.70 |
1.26 |
1.50 |
|
|
Human GI absorption (% absorbed) |
86.84 |
100 |
91.53 |
|
|
Skin Permeability (log Kp) |
-2.70 |
-3.18 |
-1.51 |
|
|
P-glycoprotein substrate |
No |
No |
No |
|
|
P-glycoprotein I inhibitor |
No |
No |
No |
|
|
P-glycoprotein II inhibitor |
No |
No |
No |
|
|
Distribution |
Human volume of distribution (log VDss) |
0.27 |
-0.38 |
0.36 |
|
Fraction unbound |
0.36 |
0.56 |
0.21 |
|
|
BBB permeability (log BB) |
-0.28 |
-0.22 |
0.66 |
|
|
CNS permeability (log PS) |
-0.33 |
-2.93 |
-1.9 |
|
|
Metabolism |
CYP2D6 substrate |
No |
No |
No |
|
CYP3A4 substrate |
No |
No |
No |
|
|
CYP1A2 inhibitor |
No |
No |
No |
|
|
CYP2C19 inhibitor |
No |
No |
No |
|
|
CYP2C9 inhibitor |
No |
No |
No |
|
|
CYP2D6 inhibitor |
No |
No |
No |
|
|
CYP3A4 inhibitor |
No |
No |
No |
|
|
Excretion |
Total Clearance (log ml/min/kg) |
0.76 |
0.58 |
1.75 |
|
Renal OCT2 substrate |
No |
No |
No |
|
|
Toxicity |
Max. human tolerated dose (log mg/kg/day) |
0.07 |
0.69 |
0.10 |
|
hERG I inhibitor |
No |
No |
No |
|
|
hERG II inhibitor |
No |
No |
No |
|
|
Oral Rat Acute Toxicity (LD50) (mol/kg) |
3.18 |
2.42 |
1.56 |
|
|
Hepatotoxicity |
Yes |
Yes |
No |
|
|
Skin Sensation |
No |
No |
Yes |
|
|
Absorption |
Water solubility (log mol/L) |
-3.81 |
-1.72 |
-5.39 |
|
Caco2 permeability (log Papp in 10 cm/s) |
1.70 |
1.26 |
1.50 |
|
The toxicity prediction revealed that IV had the highest maximum human tolerated dose (0.69 log mg/kg/day) followed by VII and II with 0.10 and 0.07 log mg/kg/day, respectively. Moreover, none of the compounds were hERG I and II inhibitors. The rat oral toxicity of II (3.18 mol/kg) was higher than both IV (2.42 mol/kg) and VII (1.56 mol/kg). Additionally, all the compounds were hepatotoxic except VII which also exhibited skin sensation.
The oral bioavailability radar of the top-docked compounds is displayed in Figure 9, revealing the lipophilicity (LIPO), size, polarity (POLAR), insolubility (INSOLU), insaturation (INSATU), and flexibility (FLEX). Only compound II demonstrated a stretch outside the acceptable bioavailability radar, stretching more toward flexibility. Although both IV and VII were within the acceptable range, IV was stretched towards insaturation while VII was towards flexibility.
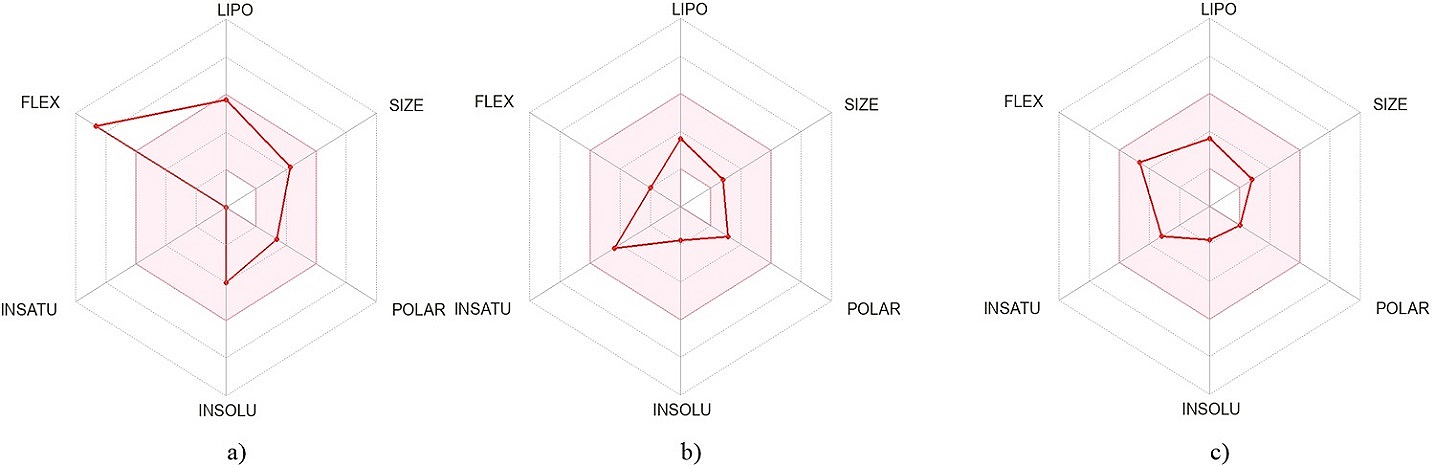
Figure 9. Oral bioavailability radar of the top-docked compounds derived from the LIPO, size, POLAR, INSOLU, INSATU, and FLEX. Note: a) Compound II, b) Compound IV, and c) Compound VII.
Discussion
The therapeutic roles of plant-based drugs are closely associated with their phytochemical components present (36). In the present study, saponins and terpenoids were detected though alkaloids, steroids, glycosides, and flavonoids undetected. This might be due to the polarity of the extracting solvent as ethanol in our study might be favorable for saponins and terpenoids detected (37). These phytochemicals exert pharmacological and therapeutic effects (38, 39). Saponins were previously reported in the ethanol extract of N. excelsa though at a lower concentration (12.01± 0.01) compared to our reported value (27.33 ±1.20). This might be attributed to the solvent concentration used for extraction (40). Saponins were also reported previously in the ethanol stem bark extract of N. excelsa (17, 41). In a similar study on ethanol stem bark extract of N. excelsa, saponins were absent though glycosides and flavonoids were detected (42). In another study, saponins and terpenoids were present in the ethanol stem bark extract of N. excelsa in addition to flavonoids and alkaloids (43). The overall variation in the phytochemicals detected in our study and previous studies might be attributed to difference in both solvent (concentration inclusive) and extraction technique employed (37).
The phytoconstituents identified in our study are linked to different pharmacological activities (44). Compounds identified in our study were made up of long-chain aliphatic compounds made up of mostly fatty acids, and aromatic alcohols. In a similar study, methyl palmitate, and (9Z)-octadeca-9,17-dienal were detected which agrees with our study (45). Some fatty acids act as anti-inflammatory and antioxidant agents (46). The six double bonds of squalene make it an anti-peroxidant thus, the pharmacological property of squalene against oxidative stress can be attributed to its ability to absorb oxygen singlets during autohydrolysis processes (47). Other studies in infants demonstrated that squalene stimulates the detoxification processes in the liver by activating P450 enzymes (48). (9Z)-octadeca-9,17-dienal was reported to have an antimicrobial property (49). Palmitic acid isolated from terpene was observed to have bactericidal and fungicidal effects (50). The phytoconstituents reported in this study might work individually or synergistically to exert their antimicrobial, antidiabetic, and anti-inflammatory (51-53).
The protein targets reported in our study were previously attributed to the pathology of malaria, inflammation, and oxidative stress. In our study, the MD and MDS results depict different interactions of these targets with the identified compounds, possibly leading to a disruption of the protein function. DHFRTS is a vital enzyme for DNA synthesis in the parasite, required for folate synthesis (via the reduction of dihydrofolate to tetrahydrofolate requiring NADPH) as well as thymidylate, vital precursors for the process (54). Moreover, Asp15, Phe57, Asp53, Ile13, and Met54 found in the active site of the enzyme (55), interacted with compound II. This might translate to enzyme inhibition by preventing the binding of its substrate. Moreover, the MDS depicting high cluster movement and residue displacement might further support the activity disruption stated.
The malaria parasite energy source is targeted by inhibiting the PfHT1, a vital facilitator of glucose uptake via the parasite plasma membrane, the rate-limiting stage of the parasite glycolysis (56, 57). Thus, this enzyme presents a vital target for malaria therapy (58). In our report, the compounds were docked in the binding pocket of the enzyme’s substrate previously reported (59), interacting with the pocket residues. The most favorable interaction was exhibited by IV, interacting with the pocket residues, including HBs with Asn48 and Lys51. The binding of IV to the enzyme might hinder the attachment of its substrate, subsequently inhibiting its activity. Moreover, the MDS showed cluster mobility and residue displacement at the N- and C-terminals of the enzyme, a possible stretching of the binding pocket, and activity disruption.
The product of INOS catalysis (nitric oxide from L-arginine promotes and regulates inflammation to some extent via COX2 synthesis (60). Thus, an inflammatory response mediator. Additionally, increased INOS concentration is observed in inflammation. The interaction of this enzyme with some small molecules leading to a possible activity disruption might minimize its pro-inflammatory effect. In our study, compound VII exhibited a favorable docking interaction with the enzyme which might disrupt its activity. The MDS further supports the possibility of inhibition due to the observed high residue fluctuations within the enzyme structure. A similar observation was made in the interaction of IV with COX2. The COX enzymes catalyze the initial steps in arachidonic acid and prostaglandin synthesis (61). COX2 is a proinflammatory enzyme released in response to extracellular and intracellular physiological stimuli, leading to the accumulation of prostaglandin E2 (62). Moreover, the COX2 gene is over-expressed and upregulated during inflammation.
XO is targeted in antioxidant therapy due to its key role in oxidative stress (63). XO catalyzes a key step in purine metabolism, the conversion of xanthine to hypoxanthine then uric acid. In these series of reactions, reactive oxygen species, such as peroxide and superoxide are generated. Therefore, minimizing its activity might lead to beneficial effects. In the present study, XO interacted with VII with a low BA and Ki in addition to HB formed with Asp262 and Glu263 might lead to a stable complex and disruption of enzyme activity. Moreover, the MDS showed high cluster mobility within the enzyme structure which further supports a possible inhibition of the enzyme. CytP450 21A2 activities have been associated with free radical generation (64). This enzyme generates carbon radicals catalyzing steroid synthesis from progesterone and 17α-hydroxyprogesterone hydroxylation, a key step in adrenal steroid synthesis (65, 66). MPO is an oxidoreductase microbicidal enzyme secreted by the neutrophils and macrophages as part of the immune system (67). Its activity generates free radicals from chlorides and peroxides to yield HOCl. Therefore, a target against oxidative stress.
The ADMET prediction revealed the pharmacological properties of the compounds and their possible application as drug candidates. The ADMET prediction group compounds based on solubility into poorly soluble (<-10), moderately soluble (<-6 to -10), soluble (<-4 to -6), very soluble (<-2 to -4), and highly soluble (>0) (35). Thus, in our study, all the compounds were fairly soluble however, compound IV is highly soluble which isn’t surprising considering its structure. The Caco2 permeability model is used to evaluate the permeability of drugs across the human intestinal mucosa to determine its absorption with values above 0.90 considered highly permeable (34). All the reported top-docked compounds in our study were predicted to be highly permeable and absorbable. A compound with skin permeability of log Kp > -2.5 is considered a relatively low skin permeant (34). Compound VII was the only compound predicted to be non-skin permeant among the compounds.
A higher VD value translates to drug distribution in tissues than in plasma (34). Moreover, the VDss value of log VDss < -0.15 is regarded as low while values above 0.45 are high. In our study, II and VII can be considered moderately distributed while IV is poorly distributed. Furthermore, the fraction unbound predicts the attachment of a drug to plasma proteins, affecting its efficiency to transverse membranes into tissues (34). In our study, IV showed the highest fraction of unbound while VII showed the least. The crossing of a drug across the blood-brain barrier is considered in predicting side effects and toxicities of drugs targeting the brain for their pharmacological effect (34). A logBB value > 0.3 is said to be BB permeant while < -1 is poorly permeant. Our report showed VII to be highly permeant while II and IV are poorly permeant. Moreover, the logPS value which predicts CNS permeability is considered CNS permeant if > -2 while non-permeant if < -3. Thus, all the compounds can be said to be CNS permeant.
The cytochrome P450 enzymes are involved in drug metabolism, especially the detoxification process in the liver (68). All the compounds were predicted to be neither substrates nor inhibitors of these enzymes. Furthermore, the compounds were predicted to be non-substrates of renal OCT2 though VII had the highest total clearance rate while IV had the lowest. The renal OCT2 is associated with the renal uptake and disposition of drugs, influencing the clearance rate (34). The maximum human tolerable dose defines the toxicity threshold of chemical dosage in humans (34). Compound IV was predicted to be highly tolerable compared to the others. Moreover, none of the compounds were hERG inhibitors. The hERG encodes potassium channels and their inhibition leads to ventricular arrhythmia (34). Thus, all the compounds can be said to be non-potassium channel blockers.
For a given compound, the LD50 defines the dose that causes the lowest adverse effect (34). Here, VII was predicted to have the lowest LD50 among the compounds. However, it was the only one predicted to be non-hepatotoxic and also exert skin sensation. Hepatotoxicity and skin sensation are important components considered in drug design to define the possibility of a compound causing normal functional disruption of the liver and skin adverse effects, respectively (34).
Conclusion
In our study, phytochemical profiling was carried out to identify compounds in N. excelsa with possible antimalarial activity via MD, MDS, and ADMET techniques. Consequently, 29 compounds were detected with only 7 passing the Lipinski’s rule in addition to either Ghose or Veber filters. Compounds II (buminafos), IV (methyl 6-morpholinonicotinate), and VII (farnesol) were predominantly active against the protein targets, exhibiting favorable interactions. These compounds further demonstrated good pharmacological properties via the ADMET predictions with minimal toxicity predictions. However, a significant improvement might be achieved following the structural improvement of the compounds. Conclusively, the compounds reported in our study might be significant contributors to the antimalarial, anti-inflammatory, and antioxidant activity of N. excelsa. Additionally, these compounds might be considered sources of novel drugs against malaria, inflammation, and oxidative stress.
Declarations
Ethics Statement
Not applicable.
Data Availability
The unpublished data is available upon request to the corresponding author.
Funding Information
The work was funded by the Tertiary Education Trust Fund through the Institutional-based Research Fund of Adamawa State Polytechnic Yola.
Conflict of Interest
The authors declare no conflicting interest.
References
- World Health Organisation. World Malaria Report 2022. Geneva: World Health Organization; 2022.
- United Nations Children's Fund. Malaria: UNICEF; 2024 [cited 2024 01/05/2024]. Available from: https://data.unicef.org/topic/child-health/malaria/
- Center for Disease Control and Prevention. Malaria’s Impact Worldwide: Center for Disease Control and Prevention; 2024 [cited 2024 01/05/2024]. Available from: https://www.cdc.gov/malaria/malaria_worldwide/impact.html.
- Alven S, Aderibigbe B. Combination therapy strategies for the treatment of malaria. Molecules. 2019;24(19):3601.
- Tse EG, Korsik M, Todd MH. The past, present and future of anti-malarial medicines. Malar J. 2019;18(1):93.
- Sani S, Aliyu B, Haruna M, Yahya SM, Yakasai MA, Hayatu LW, et al. Local plants and diabetes management; folkloric practices in Metropolitan Kano, Nigeria. Bayero Journal of Pure and Applied Sciences. 2019;12(1):268-273.
- Kankara SS, Ibrahim MH, Mustafa M, Go R. Ethnobotanical survey of medicinal plants used for traditional maternal healthcare in Katsina state, Nigeria. S Afr J Bot. 2015;97:165-175.
- Dahiru MM. Recent advances in the therapeutic potential phytochemicals in managing diabetes. Journal of Clinical and Basic Research. 2023;7(1):13-20.
- Dahiru MM, Abaka AM, Artimas SP. Phytochemical Analysis and Antibacterial Activity of Methanol and Ethyl Acetate Extracts of Detarium microcarpum Guill. & Perr. Biology, Medicine, & Natural Product Chemistry. 2023;12(1):281-288.
- Dahiru MM, Abaka AM, Musa N. Phytochemical Analysis, In-vitro, and In-silico Antibacterial Activity of Stembark Extract of Anogeissus leiocarpus (DC) Guill and Perr. Sciences of Pharmacy. 2023;2(3):24-41.
- Dahiru MM, Musa N, Abaka AM, Abubakar MA. Potential Antidiabetic Compounds from Anogeissus leiocarpus: Molecular Docking, Molecular Dynamic Simulation, and ADMET Studies. Borneo Journal of Pharmacy. 2023;6(3):249-277.
- Dahiru MM, Ahmadi H, Faruk MU, Aminu H, Hamman AGC. Phytochemical Analysis and Antioxidant Potential of Ethylacetate Extract of Tamarindus Indica (Tamarind) Leaves by Frap Assay. Journal of Fundamental and Applied Pharmaceutical Science. 2023;3(2):45-53.
- Dahiru MM, Nadro MS. Phytochemical Composition and Antioxidant Potential of Hyphaene thebaica Fruit. Borneo Journal of Pharmacy. 2022;5(4):325-333.
- Dahiru MM, Umar AS, Muhammad M, Fari II, Musa ZY. Phytoconstituents, Fourier-Transform Infrared Characterization, and Antioxidant Potential of Ethyl Acetate Extract of Corchorus olitorius (Malvaceae). Sciences of Phytochemistry. 2024;3(1):1-10.
- Akinwunmi OA, Olatunde OC, Adefemi S. Preliminary investigation of Nauclea latifolia ripe fruits for antioxidant and antidiabetic activities. Journal of Applied and Natural Science. 2019;11(3):6.
- Me B, Besong EE, Obu DC, Obu MSU, Djobissie SFA. Nauclea latifolia: A Medicinal, Economic and Pharmacological Review. International Journal of Plant Research. 2016;6(2):19.
- Ette EO, Ubulom EPM, Ekpenyong EC, Ekong SU, Akpan EO, Tambari DV. In vivo antiplasmodial activities of Nauclea latifolia. Asian Journal of Medical Sciences. 2015;6(3):6-11.
- Vasquez M, Zuniga M, Rodriguez A. Oxidative Stress and Pathogenesis in Malaria. Frontiers in Cellular and Infection Microbiology. 2021;11.
- Thomas DC. The phagocyte respiratory burst: Historical perspectives and recent advances. Immunol Lett. 2017;192:88-96.
- Ty MC, Zuniga M, Götz A, Kayal S, Sahu PK, Mohanty A, et al. Malaria inflammation by xanthine oxidase‐produced reactive oxygen species. EMBO Mol Med. 2019;11(8):e9903.
- Scaccabarozzi D, Deroost K, Corbett Y, Lays N, Corsetto P, Salè FO, et al. Differential induction of malaria liver pathology in mice infected with Plasmodium chabaudi AS or Plasmodium berghei NK65. Malar J. 2018;17:1-9.
- Abdullahi IN, Musa S, Emeribe AU, Muhammed M, Mustapha JO, Shuwa HA, et al. Immunological and anti-oxidant profiles of malarial children in Abuja, Nigeria. Biomedicine. 2021;11(1):41.
- Lingappan K. NF-κB in oxidative stress. Current opinion in toxicology. 2018;7:81-86.
- Percário S, Moreira DR, Gomes BAQ, Ferreira MES, Gonçalves ACM, Laurindo PSOC, et al. Oxidative stress in malaria. International journal of molecular sciences. 2012;13(12):16346-16372.
- Dahiru MM, Neksumi M. Phytochemical Profiling, Antioxidant, Antidiabetic, and ADMET Study of Diospyros mespiliformis Hochst. Ex A. DC.(Ebenaceae) Leaf. Journal of Faculty of Pharmacy of Ankara University. 2024;48(2):3-24.
- Evans WC. Trease and Evans' pharmacognosy: Elsevier Health Sciences; 2009. 608 p.
- Obadoni B, Ochuko P. Phytochemical studies and comparative efficacy of the crude extracts of some haemostatic plants in Edo and Delta States of Nigeria. Global J Pure Appl Sci. 2002;8(2):203-208.
- Indumathi C, Durgadevi G, Nithyavani S, Gayathri P. Estimation of terpenoid content and its antimicrobial property in Enicostemma litorrale. Int J ChemTech Res. 2014;6(9):4264-4267.
- Dahiru MM, Badgal EB, Musa N. Phytochemistry, GS-MS analysis, and heavy metals composition of aqueous and ethanol stem bark extracts of Ximenia americana. GSC Biological and Pharmaceutical Sciences. 2022;21(3):145-156.
- Sanner MF. Python: a programming language for software integration and development. J Mol Graph Model. 1999;17(1):57-61.
- Adasme MF, Linnemann KL, Bolz SN, Kaiser F, Salentin S, Haupt VJ, et al. PLIP 2021: Expanding the scope of the protein–ligand interaction profiler to DNA and RNA. Nucleic Acids Res. 2021;49(W1):W530-W534.
- Kurcinski M, Oleniecki T, Ciemny MP, Kuriata A, Kolinski A, Kmiecik S. CABS-flex standalone: a simulation environment for fast modeling of protein flexibility. Bioinformatics. 2019;35(4):694-695.
- Tiwari SP, Fuglebakk E, Hollup SM, Skjærven L, Cragnolini T, Grindhaug SH, et al. WEBnm@ v2.0: Web server and services for comparing protein flexibility. BMC Bioinformatics. 2014;15(1):1-12.
- Pires DEV, Blundell TL, Ascher DB. pkCSM: Predicting Small-Molecule Pharmacokinetic and Toxicity Properties Using Graph-Based Signatures. J Med Chem. 2015;58(9):4066-4072.
- Daina A, Michielin O, Zoete V. SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci Rep. 2017;7(1):42717.
- Ullah N, Zahoor M, Farhat A. A review on general introduction to medicinal plants, its phytochemicals and role of heavy metal and inorganic constituents. Life Science Journal. 2014;11(7s):520-527.
- Aboshora W, Lianfu Z, Dahir M, Qingran M, Qingrui S, Jing L, et al. Effect of extraction method and solvent power on polyphenol and flavonoid levels in Hyphaene thebaica L mart (Arecaceae)(Doum) fruit, and its antioxidant and antibacterial activities. Tropical Journal of Pharmaceutical Research. 2014;13(12):7.
- Tagousop CN, Tamokou J-d-D, Kengne IC, Ngnokam D, Voutquenne-Nazabadioko L. Antimicrobial activities of saponins from Melanthera elliptica and their synergistic effects with antibiotics against pathogenic phenotypes. Chem Cent J. 2018;12(1):97.
- Yang W, Chen X, Li Y, Guo S, Wang Z, Yu X. Advances in Pharmacological Activities of Terpenoids. Nat Prod Commun. 2020;15(3):1934578X20903555.
- Thouri A, Chahdoura H, El Arem A, Omri Hichri A, Ben Hassin R, Achour L. Effect of solvents extraction on phytochemical components and biological activities of Tunisian date seeds (var. Korkobbi and Arechti). BMC Complement Altern Med. 2017;17(1):248.
- Balogun ME, Nwachukwu DC, Salami SA, Besong EE, Obu DC, Djobissie SFA. Assessment of anti-ulcer efficacy of stem bark extract of Nauclea latifolia (African peach) in rats. Am J Biomed Res. 2016;4(1):13-17.
- Okafor PC, Ebiekpe VE. Inhibitive action of ethanol extracts from Nauclea latifolia on the corrosion of mild steel in H2SO4 solutions and their. Arabian journal of chemistry. 2013;6:285-293.
- Olubodun SO, Osagie OA. Anti-inflammatory effects Ofenantia chloranthia and Nauclea latifolia on crude oil-induced oxidative stress in Albino Wistar rats. Journal of Medicine and Biomedical Research. 2018;17(1-2):53-62.
- Haudecoeur R, Peuchmaur M, Pérès B, Rome M, Taïwe GS, Boumendjel A, et al. Traditional uses, phytochemistry and pharmacological properties of African Nauclea species: A review. J Ethnopharmacol. 2018;212:30.
- Yakubu O, Schetinger M, Arowora K, Shaibu C. GC-MS Characterization and Antioxidant Properties of Partially Purified Ethanol Extract of Nauclea latifolia (African Peach) Stem Bark. Biotechnology. 2022;21:146-155.
- Aparna V, Dileep KV, Mandal PK, Karthe P, Sadasivan C, Haridas M. Anti‐inflammatory property of n‐hexadecanoic acid: structural evidence and kinetic assessment. Chem Biol Drug Des. 2012;80(3):6.
- Pham DM, Boussouira B, Moyal D, Nguyen QL. Oxidization of squalene, a human skin lipid: a new and reliable marker of environmental pollution studies. International journal of cosmetic science. 2015;37(4):8.
- Ronco AL, De Stéfani E. Squalene: a multi-task link in the crossroads of cancer and aging. Functional Foods in Health and Disease. 2013;3(12):15.
- Rajeswari G, Murugan M, Mohan VR. GC-MS analysis of bioactive components of Hugonia mystax L.(Linaceae). Research Journal of Pharmaceutical, Biological and chemical sciences. 2012;3(4):301-308.
- Popova MP, Chinou IB, Marekov IN, Bankova VS. Terpenes with antimicrobial activity from Cretan propolis. Phytochemistry. 2009;70(10):10.
- Uduwana S, Abeynayake N, Wickramasinghe I. Synergistic, antagonistic, and additive effects on the resultant antioxidant activity in infusions of green tea with bee honey and Citrus limonum extract as additives. Journal of Agriculture and Food Research. 2023;12:100571.
- Ayaz M, Ullah F, Sadiq A, Ullah F, Ovais M, Ahmed J, et al. Synergistic interactions of phytochemicals with antimicrobial agents: Potential strategy to counteract drug resistance. Chemico-Biological Interactions. 2019;308:294-303.
- Zhang L, Virgous C, Si H. Synergistic anti-inflammatory effects and mechanisms of combined phytochemicals. J Nutr Biochem. 2019;69:19-30.
- Kongsaeree P, Khongsuk P, Leartsakulpanich U, Chitnumsub P, Tarnchompoo B, Walkinshaw MD, et al. Crystal structure of dihydrofolate reductase from Plasmodium vivax: pyrimethamine displacement linked with mutation-induced resistance. Proc Natl Acad Sci. 2005;102(37):13046-13051.
- Melaku Y, Solomon M, Eswaramoorthy R, Beifuss U, Ondrus V, Mekonnen Y. Synthesis, antiplasmodial activity and in silico molecular docking study of pinocembrin and its analogs. BMC chemistry. 2022;16(1):36.
- Krishna S, Webb R, Woodrow C. Transport proteins of Plasmodium falciparum: defining the limits of metabolism. Int J Parasitol. 2001;31(12):1331-1342.
- Landfear SM. Glucose transporters in parasitic protozoa. Membrane Transporters in Drug Discovery and Development: Methods and Protocols. 2010:245-262.
- Huang J, Yuan Y, Zhao N, Pu D, Tang Q, Zhang S, et al. Orthosteric–allosteric dual inhibitors of PfHT1 as selective antimalarial agents. Proc Natl Acad Sci. 2021;118(3):e2017749118.
- Jiang X, Yuan Y, Huang J, Zhang S, Luo S, Wang N, et al. Structural basis for blocking sugar uptake into the malaria parasite Plasmodium falciparum. Cell. 2020;183(1):258-268.
- Papi S, Ahmadizar F, Hasanvand A. The role of nitric oxide in inflammation and oxidative stress. Immunopathologia Persa. 2019;5:e08-e08.
- Ferrer MD, Busquets-Cortés C, Capó X, Tejada S, Tur JA, Pons A, et al. Cyclooxygenase-2 inhibitors as a therapeutic target in inflammatory diseases. Curr Med Chem. 2019;26(18):3225-3241.
- Vishwakarma RK, Negi DS. The development of COX-1 and COX-2 inhibitors: a review. Int J Pharm Sci Res. 2020;11(8):3544.
- Singh A, Singh K, Sharma A, Kaur K, Chadha R, Singh Bedi PM. Past, Present and Future of Xanthine Oxidase Inhibitors: Design Strategies, Structural and Pharmacological Insights, Patents and Clinical Trials. RSC Medicinal Chemistry. 2023.
- Veith A, Moorthy B. Role of cytochrome P450s in the generation and metabolism of reactive oxygen species. Current Opinion in Toxicology. 2018;7:44-51.
- Pallan PS, Wang C, Lei L, Yoshimoto FK, Auchus RJ, Waterman MR, et al. Human Cytochrome P450 21A2, the Major Steroid 21-Hydroxylase: Structure of the enzyme· progesterone substrate complex and rate-limiting c–h bond cleavage. J Biol Chem. 2015;290(21):13128-13143.
- Yoshimoto FK, Zhou Y, Peng H-M, Stidd D, Yoshimoto JA, Sharma KK, et al. Minor activities and transition state properties of the human steroid hydroxylases cytochromes P450c17 and P450c21, from reactions observed with deuterium-labeled substrates. Biochemistry. 2012;51(36):7064-7077.
- Chen S, Chen H, Du Q, Shen J. Targeting Myeloperoxidase (MPO) Mediated Oxidative Stress and Inflammation for Reducing Brain Ischemia Injury: Potential Application of Natural Compounds. Front Physiol. 2020;11.
- Vrbanac J, Slauter R. Chapter 3 - ADME in Drug Discovery. In: Faqi AS, editor. A Comprehensive Guide to Toxicology in Nonclinical Drug Development (Second Edition). Boston: Academic Press; 2017. p. 39-67.
 ETFLIN
Notification
ETFLIN
Notification







