Penicillin Binding Protein Mutation and Beyond: A Comprehensive Approach to Addressing Streptococcus pneumoniae Resistance
by Jajang Japar Sodik, Yani Mulyani ★
Academic editor: Khalid Abdelsamea Mohamedahmed
Sciences of Pharmacy 2(1): 37-45 (2023); https://doi.org/10.58920/sciphar02010050
This article is licensed under the Creative Commons Attribution (CC BY) 4.0 International License.
13 Jan 2023
07 Mar 2023
21 Mar 2023
28 Mar 2023
Abstract: Antibiotic resistance is a critical issue that threatens global health. Streptococcus pneumoniae, a common respiratory pathogen, has developed resistance to β-lactam antibiotics, which is of great concern. The primary mechanism of β-lactam resistance in S. pneumoniae is the acquisition of PBP genes from related species through recombination, resulting in changes in penicillin-binding proteins that affect cell wall synthesis. This mini-review summarized the understanding of β-lactam resistance in S. pneumoniae, focusing on the mechanisms and factors influencing resistance development. We conducted a comprehensive literature search using PubMed and Google Scholar, with the keywords ‘Resistant Streptococcus pneumonia’, ‘Mechanism of Streptococcus pneumoniae resistant’, and ‘Penicillin Resistant on Binding Protein of Streptococcus pneumonia’. Our literature review revealed that the prevalence of β-lactam resistance in S. pneumoniae has increased, leading to treatment failures and mortality rates. In addition to acquiring PBP genes, mutations in other PBP and non-PBP genes can contribute to resistance. Furthermore, S. pneumoniae has intrinsic resistance to various antibiotics, including first-generation polypeptides, aminoglycosides, and quinolones. Our review highlights the importance of understanding the complex mechanisms of β-lactam resistance and the need for continued efforts to monitor and control antibiotic resistance in S. pneumoniae. Further research is needed to explore novel strategies for combating antibiotic resistance in this pathogen.
Keywords: Streptococcus pneumoniaeAntibiotic resistanceβ-lactamPenicillin-binding proteins
Introduction
The discovery of penicillin in the first decade of the twentieth century, and all subsequent antibiotics, is undoubtedly one of the most significant triumphs in medicine and pharmacology. Not only have these treatments saved millions of lives, but they are also essential to be used in frequent hospital operations such as general surgery, organ transplantation, dialysis for renal failure, and chemotherapy for cancer, where their capacity to cure secondary infections is critical. Unfortunately, antibiotics' potential to heal infectious illnesses is seriously jeopardized due to the introduction and spread of antibiotic-resistant bacteria (1).
The capacity of microorganisms to resist the action of antimicrobial agents, which happens when antibiotics lose their effectiveness in preventing bacterial growth, is referred to as antibiotic resistance (2, 3). According to the European Center for Disease Prevention and Control (ECDC), around 25,000 Europeans die each year due to resistance, and an extra €1.5 billion is spent on patient care costs (1). According to the Centers for Disease Control and Prevention (CDC), a comparable number of fatalities occur in the United States. The World Health Organization projects that rising antibiotic resistance will cause 10 million deaths by 2050 (4, 5).
Antibiotic resistance has emerged as a severe issue in the medical world. Several factors impact antibiotic resistance, including the increased use of antibiotics in illness treatment. Another concern is the proliferation of antibiotic classes, which will make it simpler for bacteria to evolve resistance to antibiotics in the future (6). The rise of Streptococcus Pneumonia resistance to β-lactam antibiotics is a particularly concerning kind of antibiotic resistance. Streptococcus pneumonia is a bacteria that has caused millions of fatalities worldwide. These bacteria are general integrants of the human nasopharyngeal microbiota, although they can move and invade sterile tissues and organs. Streptococcus pneumoniae is a leading cause of morbidity and death in lower respiratory tract infections in children under five worldwide. Between 2000 and 2015, 294,000 HIV-uninfected children aged 0 to 59 months died from pneumococci, making pneumonia the most frequent illness (7).
Penicillin receptor binding proteins impact antibiotic resistance in Streptococcus pneumoniae. Penicillin-binding protein has been generally examined and linked to antibiotic resistance in Streptococcus pneumoniae bacteria. As a result, it is critical to understand the role of penicillin-binding protein in antibiotic activity. One of the most effective therapies is the exact identification of changes in the penicillin-binding protein that impact the incidence of antibiotic resistance so that the benefit of the medication may be increased while also preventing broader dissemination. Because of the advent of widespread antibiotic resistance, analysis and study related to this topic are required to establish the kind of antibiotic resistance induced by Streptococcus pneumoniae, as well as precise and accurate detection to limit the impact of Streptococcus pneumoniae resistance. This identification allows further efforts to limit and treat the impact of Streptococcus pneumoniae infection.
Methodology
The method employed in this study was a literature review of several international journals published in PubMed, ScienceDirect, and Google Scholar in the recent ten years (2012-2022). The keywords used were ‘Resistant Streptococcus pneumonia’, ‘Mechanism of Streptococcus pneumoniae resistant’, and ‘Penicillin Resistant on Binding Protein of Streptococcus pneumonia’. The initial number of articles found was 35 articles. The articles were then yielded 13 articles (published from 2012 to 2022) that provided information regarding the prevalence of antibiotic resistance, Streptococcus pneumoniae resistance, and the resistance mechanisms.
Streptococcus pneumoniae and Antibiotics Resistance
It was found that the β-lactam class of antibiotics had the highest level of resistance to Streptococcus pneumoniae (see Table 1). The abuse of β-lactams has contributed to the rise of penicillin-resistant Streptococcus pneumoniae, a problem that persists and is considered a public health issue. Drug-resistant S. pneumoniae is a severe problem in the United States, according to a 2013 study issued by the Centers for Disease Control and Prevention (see Table 1). Antibiotic-resistant Streptococcus pneumoniae strains are estimated to cause more than 1.2 million infections yearly, resulting in more than 7000 fatalities in the United States despite medication availability (8). Penicillin's Minimum Inhibitory Concentration (MIC) has been raised throughout time, and there have been reports of penicillin-resistant strains of Streptococcus pneumoniae.
Although the clinical consequences of pneumococcal pneumonia produced by nonsusceptible and susceptible strains are not dissimilar, the use of penicillin as a treatment option is declining. CLSI distinguishes between meningitis and non-meningitis syndromes and classifies MIC breakpoints based on the route of penicillin administration (i.e., parenteral vs. oral route). Aside from drug absorption, distribution to the site of action is a pharmacokinetic characteristic that directly impacts treatment success. Penicillin resistance is reduced when the CLSI breakpoint is changed.
In recent years, bacteria have emerged as the primary cause of the onset of illnesses, indicating a reduction in the quality of human health. According to a study, Streptococcus pneumoniae is one of the bacteria that cause many of these illnesses (8).
Streptococcus pneumoniae is classified into many kinds depending on its antibiotic resistance. As a result of this resistance, handling germs becomes more challenging, and the options for therapy become restricted, lowering the quality of treatment and healthcare. Streptococcus pneumoniae was initially solely resistant to penicillin antibiotics. However, as time passes, these bacteria's resistance spreads and manifests in numerous forms, including resistance to macrolides, lincosamides, fluoroquinolones, tetracyclines, and sulfamethoxazole-trimethoprim (See Table 1) (8).
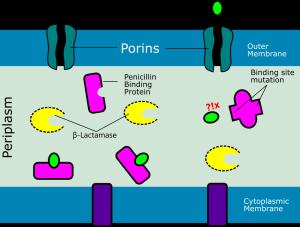
β-lactams are antibiotics responsible for inhibiting cell wall synthesis through binding to specific enzymes called Penicillin Binding Proteins (PBP) (9). Over time, the resistance level to different antimicrobial agents in Streptococcus pneumoniae increased and became a global problem. The prevalence of carrier-resistant strains has also expanded, exacerbating the problem. In addition, Streptococcus pneumoniae has intrinsic resistance to a wide range of antibiotics, including polypeptides, aminoglycosides, and first-generation quinolones (8).
β-lactam is a hydrophilic component that can enter the bacterial cell via the outer membrane's porin channels. β-lactams work by binding to penicillin-binding protein (PBP)-trans-carboxypeptidase, an enzyme involved in the production of the peptidoglycan chain of the bacterial inner membrane. PBP interaction with β-lactam antibiotics leads to peptidoglycan production inhibition, cell division halt, and cell death. The interaction of β-lactams with the active site of PBP is strongly influenced by its structure. As a result, the presence of the β-lactam ring is critical in antibiotic and antibacterial action. When β-lactam engages with PBP, an enzyme-acyl complex is produced, and the C-N bonds in the four β-lactam rings are broken (14).
Table 1. Prevalence and resistance mechanisms of Streptococcus pneumoniae to certain antibiotics.
|
Class |
Prevalence |
Resistant Mechanism |
MIC |
Ref(s) |
|
β-lactam |
Penisilin |
|
|
|
|
Penisilin: 35 % |
Modification of penicillin-binding proteins |
1 ppm |
||
|
Oxacillin : 52 % |
Modification of penicillin-binding proteins |
4 ppm |
||
|
|
||||
|
Cefalophorines |
|
|
|
|
|
Cefuroxime: 29,9% |
Modification of penicillin-binding proteins |
>4 ppm |
||
|
Ceftriaxone: 0-1% |
Modification of penicillin-binding proteins |
2 ppm |
||
|
Ceftaroline: 0-1% |
Modification of penicillin-binding proteins |
- |
||
|
|
||||
|
Imipenem: 23.4% |
Modification of penicillin-binding proteins |
1 ppm |
||
|
Macrolides |
20-40% Vancomycin: Erythromycin: |
Mutations occur in the ribosomal RNA (rRNA) binding site for the macrolide antibiotic |
1 ppm >256 ppm |
|
|
Lincosamide |
21.80% |
Modification of the ribosome binding site through mutations in the 23S rRNA gene |
- |
(8) |
|
Fluoroquinolones |
1-2% Levofloxacin: Moxifloxacin: |
Fluoroquinolones target bacterial DNA gyrase and topoisomerase IV, enzymes in DNA replication and repair. Mutations in the genes encoding these enzymes can reduce the binding affinity of the antibiotic to its target site, making it less effective. |
2 ppm 1 ppm |
|
|
Tetracycline |
36.8 % |
The ribosomal protection protein mechanism is more specific to tetracycline resistance. |
>8 ppm |
|
|
Trimethoprim-sulfamethoxazole |
29.7 % |
Trimethoprim-sulfamethoxazole works by targeting two different enzymes involved in the folate synthesis pathway, and mutations in these enzymes can reduce the binding affinity of the antibiotics, causing resistance. |
>4 ppm |
|
The resistance mechanism to β-lactams are as follows: (i) synthesis of β-lactamases that can damage β-lactams; (ii) decrease in permeability of the bacterial outer membrane causes a decrease in porin expression; (iii) changes in PBP structure; and (iv) active release of β-lactams from the bacterial cell (efflux system). β-lactamase production is thought to be a primary mechanism in developing clinically significant resistance to β-lactam in gram-negative bacteria. Genetic mutations result in substituting numerous amino acids in the protein sequence, affecting the structure of enzymes capable of hydrolyzing different antibiotics. Mutations might occur quickly as the microorganism grows resistant to antibiotics after therapy (14). The primary mechanism of β-lactam resistance in Streptococcus pneumonia is a recombination of the PBP gene acquired from similar species (such as Streptococcus mitis and Streptococcus oralis) (9). The low degree of penicillin resistance might be mainly attributed to alterations in PBP2x and -2b, resulting in non-penicillin-susceptible Streptococcus pneumonia (PNSP). However, a high resistance level can only be achieved by combining three PBP changes PBP1a, -2b, and -2x. An illustration showing the mechanism of β-lactam resistance can be seen in Figure 1.
Since Streptococcus pneumoniae is a facultative anaerobic bacterium, the electron transport chain is incomplete, conferring a low level of natural resistance to several antibiotics (see Table 2). There are six PBPs in Streptococcus pneumonia, only three of which are associated with antibiotics resistance: PBP1a, PBP2x, and PBP2b.

Figure 1. Mechanism of beta-lactam antibiotic resistance by mutation of penicillin-binding proteins (PBPs). This illustration was created inspired by the work of Gian MR. et al. (15).
Table 2. Relation of protein mutation in Streptococcus pneumoniae and β-lactams.
|
PBP |
Mutation |
Resistant Antibiotics |
|
PBP2x |
Thr550Ala |
Cefotaxime |
|
T338A |
Penicillin |
|
|
PBP2b |
Thr446Ala |
Piperacillin |
|
T451A |
Penicillin |
|
|
PBP1a |
Mosaic gene |
Penicillin and cephalosporin |
|
cp0A |
Gly12Val |
Piperacillin |
|
ciaH |
Thr230Pro |
Cefotaxime |
|
Ala203Val |
- |
|
|
MurM |
Mosaic gene |
Penicillin and cephalosporin |
Mouz N. et al. (1999) reported that it has been shown that positions 338 and 571 are important determinants for Streptococcus pneumoniae resistance to beta-lactam antibiotics (16). Among a series of 25 PBP2x sequences from clinical isolates, three contained T338A and Q552E mutations, and one showed a combination of T338A, Q552E, and S571P substitutions. The double mutant S-PBP2x*T338A, Q552E showed more than 90 and 80% decrease in acylation efficiency for cefotaxime and Pen G, respectively, compared to S-PBP2x. The T550A mutation occurs specifically because it causes high-level cefotaxime resistance and penicillin hypersensitivity simultaneously in the PBP2x mosaic gene or even as a single mutation in pbp2x from clinical isolates. The second substitution in the same codon, T550G, further increases cefotaxime resistance (17, 18). The 550A mutation leads to a 20-fold decrease in acylation efficiency for cefotaxime, possibly due to the deletion of hydrogen bonds between T550 and the carboxylate moiety attached to the six-membered ring of second and third-generation cephalosporins (16, 18).
In PBP 2b, the adjacent Thr446Ala with homolog Ser443SerAsn is a mutation that occurs with piperacillin and is described in all clinical isolates with low-affinity PBP 2b but differs from point mutations found in laboratory mutants. The mutated PBP 2b also significantly reduces the divisional response to piperacillin, indicating that this mutation is important in developing resistance in clinical isolates (16). The T446A amino acid substitution in PBP2b is known to reduce the binding affinity for penicillin by 60%, as seen in the study isolates with penicillin MIC >0.25 mg/mL (19).
Mutations in the penicillin-binding protein 1a (pbp1a) region increase the minimal inhibitory concentration (MICs) of penicillin and cefotaxime to >0.5 mg/mL, with substitutions at residues T371A and TSQF (574-577) NTGY being important for increasing penicillin resistance (20, 21). Another recent genome-wide association discovered 301 single-nucleotide polymorphisms, 73 of which induce amino acid alterations in cell wall production genes. Furthermore, the proteins MurM and MurN, expressed by the murMN operon, cause irregular cell wall formation by replacing linear muropeptides with atypically branched ones linked to penicillin resistance (22). Although MurM alone is insufficient to generate penicillin resistance, it is critical in achieving the greatest levels of penicillin and cephalosporin resistance.
Combating Streptococcus pneumoniae Resistance
Overcoming resistance to Streptococcus pneumoniae is a major challenge in treating infectious diseases. One approach to address this issue is the development of new antibiotics with more potent inhibitory activity against S. pneumoniae. In the last five years, the FDA has approved several new antibiotics for treating S. pneumoniae infections, including Omadacycline, Lefamulin, Eravacycline, Delafloxacin, Plazomicin, and Cefiderocol. Omadacycline and Lefamulin are tetracycline and pleuromutilin antibiotics effective in treating pneumonia and skin and soft tissue infections caused by S. pneumoniae resistant to other antibiotics (23-25). Eravacycline, a newer generation tetracycline antibiotic, is approved for treating intra-abdominal and urinary tract infections caused by antibiotic-resistant bacteria, including S. pneumoniae (26, 27). Delafloxacin is a new fluoroquinolone antibiotic effective in treating skin and soft tissue infections caused by S. pneumoniae (28). Plazomicin is a new aminoglycoside antibiotic approved for treating urinary tract infections caused by antibiotic-resistant bacteria, including S. pneumoniae (29). Cefiderocol, a third-generation cephalosporin antibiotic, is approved for treating difficult-to-treat infections caused by antibiotic-resistant bacteria, including S. pneumoniae (30). Proper use of antibiotics can help slow the development of antibiotic resistance and maintain the effectiveness of these drugs in treating infectious diseases in the future.
Vaccination represents a crucial aspect of the efforts to prevent Streptococcus pneumoniae infections. Vaccines have been proven to significantly reduce the incidence of infections caused by S. pneumoniae, which can lead to a decreased likelihood of the emergence of resistant strains. Three types of pneumococcal vaccines are currently available, including pneumococcal polysaccharide vaccine (PPV), pneumococcal conjugate vaccine (PCV), and pneumococcal polysaccharide-conjugate vaccine (PCV-P) (31, 32). PCV and PCV-P vaccines are designed to elicit a better immune response than PPV. PCV vaccines are available in various formulations, depending on the number of pneumococcal serotypes captured in the vaccine (33). PCV13 is widely used in high-risk children and adults, while PCV10 is used in some countries (34, 35). Studies have shown that the use of pneumococcal conjugate vaccine has significantly reduced the number of pneumococcal infections, including infections caused by serotypes included in the vaccine (36, 37). In children, PCV13 has been demonstrated to be effective in reducing the incidence of pneumococcal invasive and non-invasive diseases, such as pneumonia, otitis media, and meningitis (38). In countries that have introduced the PCV vaccine in national immunization programs, a significant decrease has been observed in the incidence of pneumococcal infections and antibiotic resistance of the bacteria (39). However, pneumococcal vaccines do not guarantee protection from all pneumococcal serotypes (40, 41), and developing more effective and comprehensive vaccines remains a research goal.
The inappropriate use of antibiotics is a major driver of antibiotic resistance, including S. pneumoniae resistance. Overuse and misuse of antibiotics can lead to resistance, making infections more difficult to treat (42). Therefore, antibiotic stewardship is critical for preventing and controlling antibiotic resistance (43). Various steps have been taken to implement antibiotic stewardship programs to address the issue of antibiotic resistance. These include educating healthcare providers and patients on the appropriate use of antibiotics, promoting guidelines and best practices for prescribing antibiotics, and establishing surveillance systems to monitor the emergence of antibiotic resistance (44-47). Additionally, various regulations and policies have been implemented to encourage the appropriate use of antibiotics, such as guidelines for the appropriate use of antibiotics in different settings, restrictions on the use of certain antibiotics, and requirements for reporting antibiotic use and resistance data (48). One example of a policy that promotes antibiotic stewardship is the CDC's Core Elements of Hospital Antibiotic Stewardship Programs, which provides a framework for healthcare facilities to develop and implement effective antibiotic stewardship programs (49). Similarly, the World Health Organization (WHO) has developed a global action plan to combat antimicrobial resistance, which includes measures to promote the appropriate use of antibiotics and support the development of new antibiotics (50).
Author Perspective
Our mini-review highlights the critical role of penicillin-binding proteins (PBPs) mutations in the emergence and spread of antibiotic resistance in Streptococcus pneumoniae, particularly in the context of β-lactam antibiotics. Although several mechanisms of β-lactam resistance have been identified, including altered expression of efflux pumps and β-lactamases, PBPs mutations remain a major driver of resistance in this pathogen. However, much remains to be learned about the precise nature of these mutations and how they interact with other genetic and environmental factors to shape the evolution of resistance. Further research is needed to fully understand the underlying mechanisms of PBPs mutations and how they contribute to the development of resistance. Moreover, there is a need to explore novel strategies for combating antibiotic resistance in this pathogen, particularly those that target the PBPs mutations. There are several efforts have been proposed to overcome this issue. To combat resistance, novel strategies such as targeting alternative penicillin-binding proteins (51), utilizing combination therapies (52-54), and targeting biofilm formation have been proposed (55). Furthermore, alternative treatments such as vaccines, bacteriophages, and probiotics could provide a promising alternative to traditional antibiotics (56). The development of new antimicrobial agents and approaches, combined with efforts to minimize the inappropriate use of antibiotics, will be crucial in the fight against antibiotic resistance in S. pneumoniae and other bacterial pathogens. It is important to recognize that antibiotic resistance is a global issue requiring coordinated effort across the scientific, medical, and public health communities.
Conclusion
Changes in Penicillin Binding Proteins involved in the production of the cell wall of Streptococcus Pneumonia bacteria are the key factor in the emergence of antibiotic resistance, especially for β-lactam antibiotics where mutations lead to resistance in PBP1a, PBP2x, and PBP2b. The development of resistance to β-lactam antibiotics is a complex mechanism that mutations can influence in other PBP and non-PBP genes. However, the main limitation of the review is that substitutions outside the specific area of PBP genes were not examined, which may also contribute to resistance and other mechanisms.
In conclusion, understanding the underlying mechanisms of PBPs mutations and their contribution to antibiotic resistance in S. pneumoniae is crucial to developing novel strategies to combat this issue. While combining new β-lactam antibiotics with novel β-lactamase inhibitors and targeting PBP3 with new antibiotics have been proposed as effective ways to overcome resistance, alternative treatments like vaccines, bacteriophages, and probiotics may provide a promising alternative to traditional antibiotics. Additionally, combining different antibiotics or antibiotic classes and targeting biofilm formation may be effective ways to enhance the efficacy of antibiotics and overcome resistance. Further research is needed to develop effective therapies against S. pneumoniae infections.
Declarations
Ethics Statement
Not applicable.
Data Availability
Not applicable.
Funding Information
Not applicable.
Conflict of Interest
The authors declare no conflicting interest.
References
- González-Bello C. Antibiotic adjuvants – A strategy to unlock bacterial resistance to antibiotics. Bioorg Med Chem Lett. 2017 Sep;27(18):4221–8.
- Nadeem SF, Gohar UF, Tahir SF, Mukhtar H, Pornpukdeewattana S, Nukthamna P, et al. Antimicrobial resistance: more than 70 years of war between humans and bacteria. Crit Rev Microbiol. 2020 Sep 2;46(5):578–99.
- Beceiro A, Tomás M, Bou G. Antimicrobial Resistance and Virulence: a Successful or Deleterious Association in the Bacterial World? Clin Microbiol Rev. 2013 Apr;26(2):185–230.
- de Kraker MEA, Stewardson AJ, Harbarth S. Will 10 Million People Die a Year due to Antimicrobial Resistance by 2050? PLOS Med. 2016 Nov 29;13(11):e1002184.
- Pulingam T, Thong KL, Appaturi JN, Nordin NI, Dinshaw IJ, Lai CW, et al. Synergistic antibacterial actions of graphene oxide and antibiotics towards bacteria and the toxicological effects of graphene oxide on human epidermal keratinocytes. Eur J Pharm Sci. 2020 Jan;142:105087.
- Levin-Reisman I, Ronin I, Gefen O, Braniss I, Shoresh N, Balaban NQ. Antibiotic tolerance facilitates the evolution of resistance. Science (80- ). 2017 Feb 24;355(6327):826–30.
- Wahl B, O’Brien KL, Greenbaum A, Majumder A, Liu L, Chu Y, et al. Burden of Streptococcus pneumoniae and Haemophilus influenzae type b disease in children in the era of conjugate vaccines: global, regional, and national estimates for 2000–15. Lancet Glob Heal. 2018 Jul;6(7):e744–57.
- Cherazard R, Epstein M, Doan T-L, Salim T, Bharti S, Smith MA. Antimicrobial Resistant Streptococcus pneumoniae: Prevalence, Mechanisms, and Clinical Implications. Am J Ther. 2017 May;24(3):e361–9.
- Jensen A, Valdórsson O, Frimodt-Møller N, Hollingshead S, Kilian M. Commensal Streptococci Serve as a Reservoir for β-Lactam Resistance Genes in Streptococcus pneumoniae. Antimicrob Agents Chemother. 2015 Jun;59(6):3529–40.
- Salsabila K, Paramaiswari WT, Amalia H, Ruyani A, Tafroji W, Winarti Y, et al. Nasopharyngeal carriage rate, serotype distribution, and antimicrobial susceptibility profile of Streptococcus pneumoniae isolated from children under five years old in Kotabaru, South Kalimantan, Indonesia. J Microbiol Immunol Infect. 2022;55(3):482–8.
- Chambers HF. Penicillin-binding protein-mediated resistance in pneumococci and staphylococci. J Infect Dis. 1999;179(SUPPL. 2):353–9.
- Bao Y, Wang Q, Yao K, Xie G, Gao W, Huang L, et al. The changing phenotypes and genotypes of invasive pneumococcal isolates from children in Shenzhen during 2013–2017. Vaccine. 2019;37(49):7248–55.
- Zhou X, Liu J, Zhang Z, Cui B, Wang Y, Zhang Y, et al. Characterization of Streptococcus pneumoniae Macrolide Resistance and Its Mechanism in Northeast China over a 20-Year Period. Microbiol Spectr. 2022;10(5).
- Buynak JD. Understanding the longevity of the β-lactam antibiotics and of antibiotic/β-lactamase inhibitor combinations. Biochem Pharmacol. 2006;71(7):930–40.
- Rossolini GM, Arena F, Giani T. Mechanisms of Antibacterial Resistance. In: Infectious Diseases. Elsevier; 2017. p. 1181-1196.e1.
- Mouz N, Di Guilmi AM, Gordon E, Hakenbeck R, Dideberg O, Vernet T. Mutations in the active site of penicillin-binding protein PBP2x from Streptococcus pneumoniae. Role in the specificity for β-lactam antibiotics. J Biol Chem. 1999;274(27):19175–80.
- Grebe T, Hakenbeck R. Penicillin-binding proteins 2b and 2x of Streptococcus pneumoniae are primary resistance determinants for difierent classes of β-lactam antibiotics. Antimicrob Agents Chemother. 1996;40(4):829–34.
- Maurer P, Koch B, Zerfaß I, Krauß J, van der Linden M, Frère JM, et al. Penicillin-binding Protein 2x of Streptococcus pneumoniae: Three New Mutational Pathways for Remodelling an Essential Enzyme into a Resistance Determinant. J Mol Biol. 2008;376(5):1403–16.
- Varghese R, Neeravi A, Subramanian N, Baskar P, Anandhan K, Veeraraghavan B. Analysis of Amino Acid Sequences of Penicillin-Binding Proteins 1a, 2b, and 2x in Invasive Streptococcus pneumoniae Nonsusceptible to Penicillin Isolated from Children in India. Microb Drug Resist. 2021;27(3):311–9.
- Ahmadi A, Yaghoubi S, Irajian G. Molecular Analysis of PBP1A in Streptococcus pneumoniae Isolated from Clinical and Normal Flora Samples in Tehran, Iran: A Multicenter Study. Microb Drug Resist. 2019;25(1):39–46.
- Job V, Carapito R, Vernet T, Dessen A, Zapun A. Common alterations in PBP1a from resistant Streptococcus pneumoniae decrease its reactivity toward β-lactams: Structural insights. J Biol Chem. 2008;283(8):4886–94.
- Chewapreecha C, Marttinen P, Croucher NJ, Salter SJ, Harris SR, Mather AE, et al. Comprehensive Identification of Single Nucleotide Polymorphisms Associated with Beta-lactam Resistance within Pneumococcal Mosaic Genes. Guttman DS, editor. PLoS Genet. 2014 Aug 7;10(8):e1004547.
- Liapikou A, Cilloniz C, Palomeque A, Torres T. Emerging antibiotics for community-acquired pneumonia. Expert Opin Emerg Drugs. 2019 Oct 2;24(4):221–31.
- Falcó V, Burgos J, Almirante B. An overview of lefamulin for the treatment of community acquired bacterial pneumonia. Expert Opin Pharmacother. 2020 Apr 12;21(6):629–36.
- Abbas M, Paul M, Huttner A. New and improved? A review of novel antibiotics for Gram-positive bacteria. Clin Microbiol Infect. 2017 Oct;23(10):697–703.
- Thaden JT, Pogue JM, Kaye KS. Role of newer and re-emerging older agents in the treatment of infections caused by carbapenem-resistant Enterobacteriaceae. Virulence. 2017 May 19;8(4):403–16.
- Bassetti M, Righi E. Eravacycline for the treatment of intra-abdominal infections. Expert Opin Investig Drugs. 2014 Nov 24;23(11):1575–84.
- Candel FJ, Peñuelas M. Delafloxacin: design, development and potential place in therapy. Drug Des Devel Ther. 2017 Mar;Volume11:881–91.
- Bilinskaya A, Linder KE, Kuti JL. Plazomicin: an intravenous aminoglycoside antibacterial for the treatment of complicated urinary tract infections. Expert Rev Anti Infect Ther. 2020 Aug 2;18(8):705–20.
- Portsmouth S, van Veenhuyzen D, Echols R, Machida M, Ferreira JCA, Ariyasu M, et al. Cefiderocol versus imipenem-cilastatin for the treatment of complicated urinary tract infections caused by Gram-negative uropathogens: a phase 2, randomised, double-blind, non-inferiority trial. Lancet Infect Dis. 2018 Dec;18(12):1319–28.
- Musher DM, Anderson R, Feldman C. The remarkable history of pneumococcal vaccination: an ongoing challenge. Pneumonia. 2022 Sep 25;14(1):5.
- Berical AC, Harris D, Dela Cruz CS, Possick JD. Pneumococcal Vaccination Strategies. An Update and Perspective. Ann Am Thorac Soc. 2016 Jun;13(6):933–44.
- Løchen A, Croucher NJ, Anderson RM. Divergent serotype replacement trends and increasing diversity in pneumococcal disease in high income settings reduce the benefit of expanding vaccine valency. Sci Rep. 2020 Nov 4;10(1):18977.
- Savulescu C, Krizova P, Lepoutre A, Mereckiene J, Vestrheim DF, Ciruela P, et al. Effect of high-valency pneumococcal conjugate vaccines on invasive pneumococcal disease in children in SpIDnet countries: an observational multicentre study. Lancet Respir Med. 2017 Aug;5(8):648–56.
- Weinberger R, von Kries R, van der Linden M, Rieck T, Siedler A, Falkenhorst G. Invasive pneumococcal disease in children under 16 years of age: Incomplete rebound in incidence after the maximum effect of PCV13 in 2012/13 in Germany. Vaccine. 2018 Jan;36(4):572–7.
- Esposito S, Principi N. Impacts of the 13-Valent Pneumococcal Conjugate Vaccine in Children. J Immunol Res. 2015;2015:1–6.
- Fitzwater SP, Chandran A, Santosham M, Johnson HL. The Worldwide Impact of the Seven-valent Pneumococcal Conjugate Vaccine. Pediatr Infect Dis J. 2012 May;31(5):501–8.
- van Hoek AJ, Choi YH, Trotter C, Miller E, Jit M. The cost-effectiveness of a 13-valent pneumococcal conjugate vaccination for infants in England. Vaccine. 2012 Nov;30(50):7205–13.
- Klugman KP, Black S. Impact of existing vaccines in reducing antibiotic resistance: Primary and secondary effects. Proc Natl Acad Sci. 2018 Dec 18;115(51):12896–901.
- Slotved H-C, Dalby T, Hoffmann S. The effect of pneumococcal conjugate vaccines on the incidence of invasive pneumococcal disease caused by ten non-vaccine serotypes in Denmark. Vaccine. 2016 Feb;34(6):769–74.
- Mathew JL. Pneumococcal vaccination in developing countries: Where does science end and commerce begin? Vaccine. 2009 Jul;27(32):4247–51.
- Mancuso G, Midiri A, Gerace E, Biondo C. Bacterial Antibiotic Resistance: The Most Critical Pathogens. Pathogens. 2021 Oct 12;10(10):1310.
- Gyssens IC, Kern W V., Livermore DM. The role of antibiotic stewardship in limiting antibacterial resistance among hematology patients. Haematologica. 2013 Dec 1;98(12):1821–5.
- Majumder MAA, Singh K, Hilaire MG-S, Rahman S, Sa B, Haque M. Tackling Antimicrobial Resistance by promoting Antimicrobial stewardship in Medical and Allied Health Professional Curricula. Expert Rev Anti Infect Ther. 2020 Dec 1;18(12):1245–58.
- Biezen R, Roberts C, Buising K, Thursky K, Boyle D, Lau P, et al. How do general practitioners access guidelines and utilise electronic medical records to make clinical decisions on antibiotic use? Results from an Australian qualitative study. BMJ Open. 2019 Aug 5;9(8):e028329.
- Heil EL, Kuti JL, Bearden DT, Gallagher JC. The Essential Role of Pharmacists in Antimicrobial Stewardship. Infect Control Hosp Epidemiol. 2016 Jul 13;37(7):753–4.
- Machowska A, Stålsby Lundborg C. Drivers of Irrational Use of Antibiotics in Europe. Int J Environ Res Public Health. 2018 Dec 23;16(1):27.
- Jacobs TG, Robertson J, van den Ham HA, Iwamoto K, Bak Pedersen H, Mantel-Teeuwisse AK. Assessing the impact of law enforcement to reduce over-the-counter (OTC) sales of antibiotics in low- and middle-income countries; a systematic literature review. BMC Health Serv Res. 2019 Dec 31;19(1):536.
- CDC. Core elements of hospital antibiotic stewardship programs. Antibiot use/CDC. 2019.
- Gandra S, Alvarez-Uria G, Turner P, Joshi J, Limmathurotsakul D, van Doorn HR. Antimicrobial Resistance Surveillance in Low- and Middle-Income Countries: Progress and Challenges in Eight South Asian and Southeast Asian Countries. Clin Microbiol Rev. 2020 Jun 17;33(3).
- Selakovitch-Chenu L, Seroude L, Michel Sicard A. The role of penicillin-binding protein 3 (PBP 3) in cefotaxime resistance in Streptococcus pneumoniae. Mol Gen Genet MGG. 1993 May;239(1–2):77–80.
- Bush K. Past and Present Perspectives on β-Lactamases. Antimicrob Agents Chemother. 2018 Oct;62(10).
- Ito A, Ishida T, Tachibana H, Nakanishi Y, Tokioka F, Yamazaki A, et al. Usefulness of β-lactam and macrolide combination therapy for treating community-acquired pneumonia patients hospitalized in the intensive care unit: Propensity score analysis of a prospective cohort study. J Infect Chemother. 2021 Oct;27(10):1447–53.
- Vardakas KZ, Trigkidis KK, Falagas ME. Fluoroquinolones or macrolides in combination with β-lactams in adult patients hospitalized with community acquired pneumonia: a systematic review and meta-analysis. Clin Microbiol Infect. 2017 Apr;23(4):234–41.
- Liu Y, Huang Y, Fan C, Chi Z, Bai M, Sun L, et al. Ursolic Acid Targets Glucosyltransferase and Inhibits Its Activity to Prevent Streptococcus mutans Biofilm Formation. Front Microbiol. 2021 Sep 27;12.
- Konwar AN, Hazarika SN, Bharadwaj P, Thakur D. Emerging Non-Traditional Approaches to Combat Antibiotic Resistance. Curr Microbiol. 2022 Nov 25;79(11):330.
 ETFLIN
Notification
ETFLIN
Notification